Saccharomyces cerevisiae is permissive for replication of bovine papillomavirus type 1
- PMID: 12414966
- PMCID: PMC136905
- DOI: 10.1128/jvi.76.23.12265-12273.2002
Saccharomyces cerevisiae is permissive for replication of bovine papillomavirus type 1
Abstract
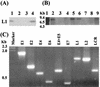
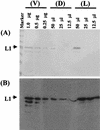
We recently demonstrated that Saccharomyces cerevisiae protoplasts can take up bovine papillomavirus type 1 (BPV1) virions and that viral episomal DNA is replicated after uptake. Here we demonstrate that BPV virus-like particles are assembled in infected S. cerevisiae cultures from newly synthesized capsid proteins and also package newly synthesized DNA, including full-length and truncated viral DNA and S. cerevisiae-derived DNA. Virus particles prepared in S. cerevisiae are able to convey packaged DNA to Cos1 cells and to transform C127 cells. Infectivity was blocked by antisera to BPV1 L1 but not antisera to BPV1 E4. We conclude that S. cerevisiae is permissive for the replication of BPV1 virus.
Figures





Similar articles
-
Saccharomyces cerevisiae: a useful model host to study fundamental biology of viral replication.Virus Res. 2006 Sep;120(1-2):49-56. doi: 10.1016/j.virusres.2005.11.018. Epub 2006 May 15. Virus Res. 2006. PMID: 16698107 Free PMC article. Review.
-
BPV1 E2 protein enhances packaging of full-length plasmid DNA in BPV1 pseudovirions.Virology. 2000 Jul 5;272(2):382-93. doi: 10.1006/viro.2000.0348. Virology. 2000. PMID: 10873782
-
Replication of bovine papillomavirus type 1 (BPV-1) DNA in Saccharomyces cerevisiae following infection with BPV-1 virions.J Virol. 2002 Apr;76(7):3359-64. doi: 10.1128/jvi.76.7.3359-3364.2002. J Virol. 2002. PMID: 11884561 Free PMC article.
-
In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype.J Virol. 1996 Sep;70(9):5875-83. doi: 10.1128/JVI.70.9.5875-5883.1996. J Virol. 1996. PMID: 8709207 Free PMC article.
-
Initiation of viral DNA replication.Adv Virus Res. 1988;34:1-42. doi: 10.1016/s0065-3527(08)60514-x. Adv Virus Res. 1988. PMID: 2843015 Review. No abstract available.
Cited by
-
Generalized substitution of isoencoding codons shortens the duration of papillomavirus L1 protein expression in transiently gene-transfected keratinocytes due to cell differentiation.Nucleic Acids Res. 2007;35(14):4820-32. doi: 10.1093/nar/gkm496. Epub 2007 Jul 9. Nucleic Acids Res. 2007. PMID: 17621583 Free PMC article.
-
Yeast and the AIDS virus: the odd couple.J Biomed Biotechnol. 2012;2012:549020. doi: 10.1155/2012/549020. Epub 2012 Jun 17. J Biomed Biotechnol. 2012. PMID: 22778552 Free PMC article. Review.
-
An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses.Proc Natl Acad Sci U S A. 2009 May 26;106(21):8778-83. doi: 10.1073/pnas.0809105106. Epub 2009 May 7. Proc Natl Acad Sci U S A. 2009. PMID: 19423673 Free PMC article.
-
Varying efficiency of long-term replication of papillomaviruses in Saccharomyces cerevisiae.Virology. 2008 Nov 10;381(1):6-10. doi: 10.1016/j.virol.2008.08.038. Epub 2008 Oct 1. Virology. 2008. PMID: 18829061 Free PMC article.
-
Saccharomyces cerevisiae: a useful model host to study fundamental biology of viral replication.Virus Res. 2006 Sep;120(1-2):49-56. doi: 10.1016/j.virusres.2005.11.018. Epub 2006 May 15. Virus Res. 2006. PMID: 16698107 Free PMC article. Review.
References
-
- Cook, J. C., J. G. Joyce, H. A. George, L. D. Schultz, W. M. Hurni, K. U. Jansen, R. W. Hepler, C. Ip, R. S. Lowe, P. M. Keller, and E. D. Lehman. 1999. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr Purif 17:477-484. - PubMed
-
- Cullen, B. R., and W. C. Greene. 1989. Regulatory pathways governing HIV-1 replication. Cell 58:423-426. - PubMed
-
- Dostatni, N., P. F. Lambert, R. Sousa, J. Ham, P. M. Howley, and M. Yaniv. 1991. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 5:1657-1671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

