Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior
- PMID: 12417656
- PMCID: PMC6758054
- DOI: 10.1523/JNEUROSCI.22-21-09305.2002
Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior
Abstract
In Drosophila, a number of key processes such as emergence from the pupal case, locomotor activity, feeding, olfaction, and aspects of mating behavior are under circadian regulation. Although we have a basic understanding of how the molecular oscillations take place, a clear link between gene regulation and downstream biological processes is still missing. To identify clock-controlled output genes, we have used an oligonucleotide-based high-density array that interrogates gene expression changes on a whole genome level. We found genes regulating various physiological processes to be under circadian transcriptional regulation, ranging from protein stability and degradation, signal transduction, heme metabolism, detoxification, and immunity. By comparing rhythmically expressed genes in the fly head and body, we found that the clock has adapted its output functions to the needs of each particular tissue, implying that tissue-specific regulation is superimposed on clock control of gene expression. Finally, taking full advantage of the fly as a model system, we have identified and characterized a cycling potassium channel protein as a key step in linking the transcriptional feedback loop to rhythmic locomotor behavior.
Figures
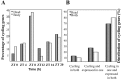

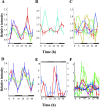
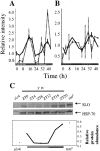

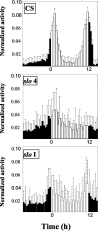
References
-
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
-
- Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
