The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats
- PMID: 12417694
- PMCID: PMC153795
- DOI: 10.1105/tpc.006155
The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats
Abstract
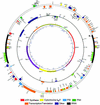
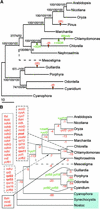
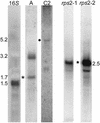
Chlamydomonas reinhardtii is a unicellular eukaryotic alga possessing a single chloroplast that is widely used as a model system for the study of photosynthetic processes. This report analyzes the surprising structural and evolutionary features of the completely sequenced 203,395-bp plastid chromosome. The genome is divided by 21.2-kb inverted repeats into two single-copy regions of approximately 80 kb and contains only 99 genes, including a full complement of tRNAs and atypical genes encoding the RNA polymerase. A remarkable feature is that >20% of the genome is repetitive DNA: the majority of intergenic regions consist of numerous classes of short dispersed repeats (SDRs), which may have structural or evolutionary significance. Among other sequenced chlorophyte plastid genomes, only that of the green alga Chlorella vulgaris appears to share this feature. The program MultiPipMaker was used to compare the genic complement of Chlamydomonas with those of other chloroplast genomes and to scan the genomes for sequence similarities and repetitive DNAs. Among the results was evidence that the SDRs were not derived from extant coding sequences, although some SDRs may have arisen from other genomic fragments. Phylogenetic reconstruction of changes in plastid genome content revealed that an accelerated rate of gene loss also characterized the Chlamydomonas/Chlorella lineage, a phenomenon that might be independent of the proliferation of SDRs. Together, our results reveal a dynamic and unusual plastid genome whose existence in a model organism will allow its features to be tested functionally.
Figures




References
-
- Adachi, J., and Hasegawa, M. (1996). MOLPHY version 2.3: Programs for molecular phylogeny based on maximum likelihood. In Computer Science Monographs 28. (Tokyo: Institute of Statistical Mathematics).
-
- Allison, L.A. (2000). The role of sigma factors in plastid transcription. Biochimie 82, 537–548. - PubMed
-
- Backert, S., Dorfel, P., and Börner, T. (1995). Investigation of plant organellar DNAs by pulsed-field gel electrophoresis. Curr. Genet. 28, 390–399. - PubMed
-
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

