Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning
- PMID: 12417731
- PMCID: PMC134074
- DOI: 10.1128/MCB.22.23.8292-8301.2002
Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning
Abstract
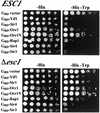
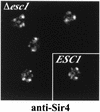
A targeted silencing screen was performed to identify yeast proteins that, when tethered to a telomere, suppress a telomeric silencing defect caused by truncation of Rap1. A previously uncharacterized protein, Esc1 (establishes silent chromatin), was recovered, in addition to well-characterized proteins Rap1, Sir1, and Rad7. Telomeric silencing was slightly decreased in Deltaesc1 mutants, but silencing of the HM loci was unaffected. On the other hand, targeted silencing by various tethered proteins was greatly weakened in Deltaesc1 mutants. Two-hybrid analysis revealed that Esc1 and Sir4 interact via a 34-amino-acid portion of Esc1 (residues 1440 to 1473) and a carboxyl-terminal domain of Sir4 known as PAD4 (residues 950 to 1262). When tethered to DNA, this Sir4 domain confers efficient partitioning to otherwise unstable plasmids and blocks the ability of bound DNA segments to rotate freely in vivo. Here, both phenomena were shown to require ESC1. Sir protein-mediated partitioning of a telomere-based plasmid also required ESC1. Fluorescence microscopy of cells expressing green fluorescent protein (GFP)-Esc1 showed that the protein localized to the nuclear periphery, a region of the nucleus known to be functionally important for silencing. GFP-Esc1 localization, however, was not entirely coincident with telomeres, the nucleolus, or nuclear pore complexes. Our data suggest that Esc1 is a component of a redundant pathway that functions to localize silencing complexes to the nuclear periphery.
Figures






References
-
- Andrulis, E. D., A. M. Neiman, D. C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394:592-595. (Erratum, 395:525.) - PubMed
-
- Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. - PubMed
-
- Buck, S. W., and D. Shore. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9:370-384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
