In silico model-driven assessment of the effects of single nucleotide polymorphisms (SNPs) on human red blood cell metabolism
- PMID: 12421755
- PMCID: PMC187553
- DOI: 10.1101/gr.329302
In silico model-driven assessment of the effects of single nucleotide polymorphisms (SNPs) on human red blood cell metabolism
Abstract
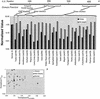
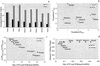
The completion of the human genome project and the construction of single nucleotide polymorphism (SNP) maps have lead to significant efforts to find SNPs that can be linked to pathophysiology. In silico models of complete biochemical reaction networks relate a cell's individual reactions to the function of the entire network. Sequence variations can in turn be related to kinetic properties of individual enzymes, thus allowing an in silico model-driven assessment of the effects of defined SNPs on overall cellular functions. This process is applied to defined SNPs in two key enzymes of human red blood cell metabolism: glucose-6-phosphate dehydrogenase and pyruvate kinase. The results demonstrate the utility of in silico models in providing insight into differences between red cell function in patients with chronic and nonchronic anemia. In silico models of complex cellular processes are thus likely to aid in defining and understanding key SNPs in human pathophysiology.
Figures


Similar articles
-
Path to facilitate the prediction of functional amino acid substitutions in red blood cell disorders--a computational approach.PLoS One. 2011;6(9):e24607. doi: 10.1371/journal.pone.0024607. Epub 2011 Sep 13. PLoS One. 2011. PMID: 21931771 Free PMC article.
-
Hemolytic anemias due to erythrocyte enzyme deficiencies.Mol Aspects Med. 1996 Apr;17(2):143-70. doi: 10.1016/0098-2997(96)88345-2. Mol Aspects Med. 1996. PMID: 8813716
-
Clinical and biochemical studies on mutant red cell enzymes mainly associated with hemolytic anemia.Jinrui Idengaku Zasshi. 1980 Jun;25(2):83-92. doi: 10.1007/BF01873607. Jinrui Idengaku Zasshi. 1980. PMID: 7431680 No abstract available.
-
[Hereditary enzyme defects of erythrocytes: glucose-6-phosphate dehydrogenase deficiency and pyruvate kinase deficiency].Ther Umsch. 2006 Jan;63(1):47-56. doi: 10.1024/0040-5930.63.1.47. Ther Umsch. 2006. PMID: 16450734 Review. German.
-
[Pathophysiology and laboratory tests of hemolytic anemia: with special reference to erythroenzymopathies].Rinsho Byori. 1989 Dec;37(12):1331-43. Rinsho Byori. 1989. PMID: 2693773 Review. Japanese.
Cited by
-
Revitalizing personalized medicine: respecting biomolecular complexities beyond gene expression.CPT Pharmacometrics Syst Pharmacol. 2014 Apr 16;3(4):e110. doi: 10.1038/psp.2014.6. CPT Pharmacometrics Syst Pharmacol. 2014. PMID: 24739991 Free PMC article.
-
Evaluation of rate law approximations in bottom-up kinetic models of metabolism.BMC Syst Biol. 2016 Jun 6;10(1):40. doi: 10.1186/s12918-016-0283-2. BMC Syst Biol. 2016. PMID: 27266508 Free PMC article.
-
Systems-biology approaches for predicting genomic evolution.Nat Rev Genet. 2011 Aug 2;12(9):591-602. doi: 10.1038/nrg3033. Nat Rev Genet. 2011. PMID: 21808261 Review.
-
Analysis of genetic variation and potential applications in genome-scale metabolic modeling.Front Bioeng Biotechnol. 2015 Feb 16;3:13. doi: 10.3389/fbioe.2015.00013. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 25763369 Free PMC article. Review.
-
Formulating genome-scale kinetic models in the post-genome era.Mol Syst Biol. 2008;4:171. doi: 10.1038/msb.2008.8. Epub 2008 Mar 4. Mol Syst Biol. 2008. PMID: 18319723 Free PMC article.
References
-
- Adediran SA. Kinetic and thermodynamic properties of two electrophoretically similar genetic variants of human erythrocyte glucose-6-phosphate dehydrogenase. Biochimie. 1996;78:165–170. - PubMed
-
- Atkinson DE. Cellular energy metabolism and its regulation. New York: Academic Press; 1977.
-
- Au SW, Gover S, Lam V, Adams MJ. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP molecule and provides insights into enzyme deficiency. Structure. 2000;8:293–303. - PubMed
-
- Betke KBE, Brewer GJ, Kirkman HN, Luzzatto L, Motulsky AG, Ramot B, Siniscalco M. Standardization of procedures for the study of glucose-6-phosphate dehydrogenase. Report of a WHO scientific group. WHO Tech Rep Ser. 1967;Ser:366. - PubMed
-
- Beutler E. Red cell metabolism. Edinburgh, England; New York: Churchill Livingstone; 1986.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
