Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation
- PMID: 12422020
- PMCID: PMC137742
- DOI: 10.1073/pnas.242596699
Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation
Abstract
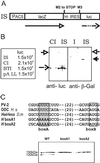
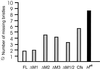
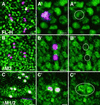
The Notch-signaling pathway controls cellular differentiation, including proliferation and cell death in all higher metazoans (including flies and men). Signal transduction through activated Notch involves the CSL group of transcriptional regulators. Notch signals need to be tightly regulated, and in Drosophila they are antagonized by the Hairless (H) protein. H silences the activity of Notch target genes by transforming the Drosophila CSL protein, Suppressor of Hairless [Su(H)], from a transcriptional activator into a repressor while recruiting one of the corepressors dCtBP or Groucho. The H protein has a calculated molecular mass of approximately 110 kDa and contains several functional domains apart from the two small corepressor-binding domains. However, although there is no indication for alternative splicing, two Hairless protein isoforms, H(p120) and H(p150), are observed throughout development. Here, we show that the smaller isoform derives from an internal ribosome entry site (IRES) within the ORF. The IRES is active in a heterologous assay and contains an essential, conserved structural element. The two Hairless isoforms have residual activity in vivo which is, however, reduced compared to a combination of both, which implies that both protein isoforms are necessary for WT function. In larval tissues, translation of the two isoforms is cell-cycle regulated: whereas the H(p150) isoform is translated during interphase, H(p120) is enriched during mitosis. Thus, the presence of either H isoform throughout the cell cycle allows efficient inhibition of Notch-regulated cell proliferation.
Figures

Similar articles
-
In vivo analysis of internal ribosome entry at the Hairless locus by genome engineering in Drosophila.Sci Rep. 2016 Oct 7;6:34881. doi: 10.1038/srep34881. Sci Rep. 2016. PMID: 27713501 Free PMC article.
-
Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless.Genes Dev. 2002 Aug 1;16(15):1964-76. doi: 10.1101/gad.987402. Genes Dev. 2002. PMID: 12154126 Free PMC article.
-
Molecular analysis of the notch repressor-complex in Drosophila: characterization of potential hairless binding sites on suppressor of hairless.PLoS One. 2011;6(11):e27986. doi: 10.1371/journal.pone.0027986. Epub 2011 Nov 18. PLoS One. 2011. PMID: 22125648 Free PMC article.
-
Hairless: the ignored antagonist of the Notch signalling pathway.Hereditas. 2006 Dec;143(2006):212-21. doi: 10.1111/j.2007.0018-0661.01971.x. Hereditas. 2006. PMID: 17362357 Review.
-
Role of suppressor of hairless in the delta-activated Notch signaling pathway.Perspect Dev Neurobiol. 1997;4(4):305-11. Perspect Dev Neurobiol. 1997. PMID: 9171444 Review.
Cited by
-
An internal ribosome entry site element directs the synthesis of the 80 kDa isoforms of protein 4.1R.BMC Biol. 2008 Dec 4;6:51. doi: 10.1186/1741-7007-6-51. BMC Biol. 2008. PMID: 19055807 Free PMC article.
-
A Small Molecule That Promotes Cellular Senescence Prevents Fibrogenesis and Tumorigenesis.Int J Mol Sci. 2022 Jun 20;23(12):6852. doi: 10.3390/ijms23126852. Int J Mol Sci. 2022. PMID: 35743290 Free PMC article.
-
Structural and functional analysis of the repressor complex in the Notch signaling pathway of Drosophila melanogaster.Mol Biol Cell. 2011 Sep;22(17):3242-52. doi: 10.1091/mbc.E11-05-0420. Epub 2011 Jul 7. Mol Biol Cell. 2011. PMID: 21737682 Free PMC article.
-
The Notch repressor complex in Drosophila: in vivo analysis of Hairless mutants using overexpression experiments.Dev Genes Evol. 2019 Jan;229(1):13-24. doi: 10.1007/s00427-018-00624-2. Epub 2019 Jan 5. Dev Genes Evol. 2019. PMID: 30612166
-
Genetic and Molecular Interactions between HΔCT, a Novel Allele of the Notch Antagonist Hairless, and the Histone Chaperone Asf1 in Drosophila melanogaster.Genes (Basel). 2023 Jan 13;14(1):205. doi: 10.3390/genes14010205. Genes (Basel). 2023. PMID: 36672946 Free PMC article.
References
-
- Artavanis-Tsakonas S., Rand, M. D. & Lake, R. J. (1999) Science 284 770-776. - PubMed
-
- Brou C., Logeat, F., Lecourtois, M., Vandekerckhove, J., Kourilsky, P., Schweisguth, F. & Israël, A. (1994) Genes Dev. 8 2491-2503. - PubMed
-
- Morel V., Lecourtois, M., Massiani, O., Maier, D., Preiss, A. & Schweisguth, F. (2001) Curr. Biol. 11 789-792. - PubMed
-
- Maier D., Nagel, A. C., Johannes, B. & Preiss, A. (1999) Mech. Dev. 89 195-199. - PubMed
-
- Bang A. G. & Posakony, J. W. (1992) Genes Dev. 6 1752-1769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

