vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405
- PMID: 12426332
- PMCID: PMC135418
- DOI: 10.1128/JB.184.23.6457-6464.2002
vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405
Abstract
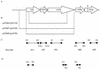
Acquired VanE-type resistance to low levels of vancomycin (MIC = 16 microg/ml) in Enterococcus faecalis BM4405 is due to the inducible synthesis of peptidoglyean precursors terminating in D-alanine-D-serine (Fines,M., B. Prichon, P. Reynolds, D. Sahm, and P. Courvalin, Antimicrob. Agents Chemother. 43:2161-2164, 1999). A chromosomal location was assigned to the vanE operon by pulsed-field gel electrophoresis and hybridization, and its sequence was determined. Three genes, encoding the VanE ligase, the VanXYE DD-peptidase, and the VanTE serine racemase, that displayed 43 to 53% identity with the corresponding genes in the vanC operon were found. In addition, two genes coding for a two-component regulatory system, VanRE-VanSE, exhibiting 60 and 44% identity with VanR,-VanS, were present downstream from vanTE. However, because of a stop codon at position 78, VanSE was probably not functional. The five genes, with the same orientation, were shown to be cotranscribed by Northern analysis and reverse transcription-PCR. The vanE, vanXYE, and vanTE genes conferred inducible low-level resistance to vancomycin after cloning in E. faecalis JH2-2, probably following cross talk with a two-component regulatory system of the host.
Figures



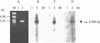

References
-
- Arias, C., M. Martín-Martinez, T. Blundell, M. Arthur, P. Courvalin, and P. Reynolds. 1999. Characterization and modelling of VanT: a novel membrane-bound serine racemase from vancomycin resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 31:1653-1654. - PubMed
-
- Arthur, M., C. Molinas, and P. Courvalin. 1992. Sequence of the vanY gene required for production of a vancomycin-inducible d,d-carboxypeptidase in Enterococcus faecium BM4147. Gene 120:11-114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

