Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p
- PMID: 12426383
- PMCID: PMC137200
- DOI: 10.1093/emboj/cdf608
Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p
Abstract
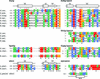
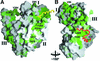
Cytosolic Sec1/munc18-like proteins (SM proteins) are recruited to membrane fusion sites by interaction with syntaxin-type SNARE proteins, constituting indispensable positive regulators of intracellular membrane fusion. Here we present the crystal structure of the yeast SM protein Sly1p in complex with a short N-terminal peptide derived from the Golgi-resident syntaxin Sed5p. Sly1p folds, similarly to neuronal Sec1, into a three-domain arch-shaped assembly, and Sed5p interacts in a helical conformation predominantly with domain I of Sly1p on the opposite site of the nSec1/syntaxin-1-binding site. Sequence conservation of the major interactions suggests that homologues of Sly1p as well as the paralogous Vps45p group bind their respective syntaxins in the same way. Furthermore, we present indirect evidence that nSec1 might be able to contact syntaxin 1 in a similar fashion. The observed Sly1p-Sed5p interaction mode therefore indicates how SM proteins can stay associated with the assembling fusion machinery in order to participate in late fusion steps.
Figures




References
-
- Bracher A. and Weissenhorn,W. (2001) Crystal structures of neuronal squid Sec1 implicate inter-domain hinge movement in the release of t-SNAREs. J. Mol. Biol., 306, 7–13. - PubMed
-
- Bracher A., Perrakis,A., Dresbach,T., Betz,H. and Weissenhorn,W. (2000) The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis. Structure Fold. Des., 8, 685–694. - PubMed
-
- Bracher A., Kadlec,J., Betz,H. and Weissenhorn,W. (2002) X-ray structure of a neuronal complexin–SNARE complex from squid. J. Biol. Chem., 277, 26517–26523. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

