Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response
- PMID: 12426388
- PMCID: PMC137192
- DOI: 10.1093/emboj/cdf600
Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response
Abstract
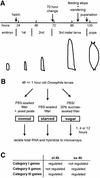
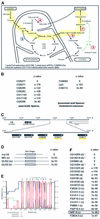
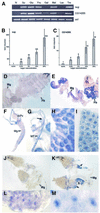
We have identified genes regulated by starvation and sugar signals in Drosophila larvae using whole-genome microarrays. Based on expression profiles in the two nutrient conditions, they were organized into different categories that reflect distinct physiological pathways mediating sugar and fat metabolism, and cell growth. In the category of genes regulated in sugar-fed, but not in starved, animals, there is an upregulation of genes encoding key enzymes of the fat biosynthesis pathway and a downregulation of genes encoding lipases. The highest and earliest activated gene upon sugar ingestion is sugarbabe, a zinc finger protein that is induced in the gut and the fat body. Identification of potential targets using microarrays suggests that sugarbabe functions to repress genes involved in dietary fat breakdown and absorption. The current analysis provides a basis for studying the genetic mechanisms underlying nutrient signalling.
Figures





References
-
- Abu-Elheiga L., Matzuk,M., Abo-Hasema,K. and Wakil,S. (2001) Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science, 291, 2613–2616. - PubMed
-
- Beadle G., Tatum,E. and Clancy,C. (1938) Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol. Bull., 75, 447–462.
-
- Blundell J. (1991) Pharmacological approaches to appetite suppression. Trends Pharm. Sci., 12, 147–157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

