Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
- PMID: 12426397
- PMCID: PMC137216
- DOI: 10.1093/emboj/cdf627
Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
Abstract
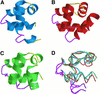
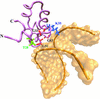
The C-terminal domain of the UvrC protein (UvrC CTD) is essential for 5' incision in the prokaryotic nucleotide excision repair process. We have determined the three-dimensional structure of the UvrC CTD using heteronuclear NMR techniques. The structure shows two helix-hairpin-helix (HhH) motifs connected by a small connector helix. The UvrC CTD is shown to mediate structure-specific DNA binding. The domain binds to a single-stranded-double-stranded junction DNA, with a strong specificity towards looped duplex DNA that contains at least six unpaired bases per loop ("bubble DNA"). Using chemical shift perturbation experiments, the DNA-binding surface is mapped to the first hairpin region encompassing the conserved glycine-valine-glycine residues followed by lysine-arginine-arginine, a positively charged surface patch and the second hairpin region consisting of glycine-isoleucine-serine. A model for the protein-DNA complex is proposed that accounts for this specificity.
Figures

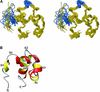
References
-
- Bonvin A.M.J.J., Houben,K., Guenneugues,M., Kaptein,R. and Boelens,R. (2001) Rapid protein fold determination using secondary chemical shifts and cross-hydrogen bond 15N–13C scalar couplings (3hbJNC′). J. Biomol. NMR, 21, 221–233. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography a NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Cavanagh J., Fairbrother, WJ., Palmer,A.G.,III and Skelton,N.J. (1996) Protein NMR Spectroscopy. Academic Press, San Diego, CA.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

