The mechanism of topoisomerase I poisoning by a camptothecin analog
- PMID: 12426403
- PMCID: PMC137726
- DOI: 10.1073/pnas.242259599
The mechanism of topoisomerase I poisoning by a camptothecin analog
Abstract
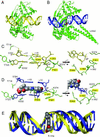
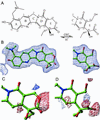
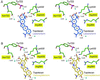
We report the x-ray crystal structure of human topoisomerase I covalently joined to double-stranded DNA and bound to the clinically approved anticancer agent Topotecan. Topotecan mimics a DNA base pair and binds at the site of DNA cleavage by intercalating between the upstream (-1) and downstream (+1) base pairs. Intercalation displaces the downstream DNA, thus preventing religation of the cleaved strand. By specifically binding to the enzyme-substrate complex, Topotecan acts as an uncompetitive inhibitor. The structure can explain several of the known structure-activity relationships of the camptothecin family of anticancer drugs and suggests that there are at least two classes of mutations that can produce a drug-resistant enzyme. The first class includes changes to residues that contribute to direct interactions with the drug, whereas a second class would alter interactions with the DNA and thereby destabilize the drug-binding site.
Figures
References
-
- Wang J. C. (1996) Annu. Rev. Biochem. 65 635-692. - PubMed
-
- Champoux J. J. (2001) Annu. Rev. Biochem. 70 369-413. - PubMed
-
- Pommier Y. (1998) Biochimie 80 255-270. - PubMed
-
- Wall M. E., Wani, M. C., Cook, C. E., Palmer, K. H., McPhail, A. T. & Sim, G. A. (1966) J. Am. Chem. Soc. 88 3888-3890.
-
- Hsiang Y. H., Hertzberg, R., Hecht, S. & Liu, L. F. (1985) J. Biol. Chem. 260 14873-14878. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

