Functional analysis of an Arabidopsis T-DNA "knockout" of the high-affinity NH4(+) transporter AtAMT1;1
- PMID: 12427993
- PMCID: PMC166647
- DOI: 10.1104/pp.102.010843
Functional analysis of an Arabidopsis T-DNA "knockout" of the high-affinity NH4(+) transporter AtAMT1;1
Abstract
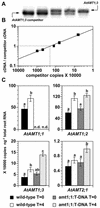
NH(4)(+) acquisition by plant roots is thought to involve members of the NH(4)(+) transporter family (AMT) found in plants, yeast, bacteria, and mammals. In Arabidopsis, there are six AMT genes of which AtAMT1;1 demonstrates the highest affinity for NH(4)(+). Ammonium influx into roots and AtAMT1;1 mRNA expression levels are highly correlated diurnally and when plant nitrogen (N) status is varied. To further investigate the involvement of AtAMT1;1 in high-affinity NH(4)(+) influx, we identified a homozygous T-DNA mutant with disrupted AtAMT1;1 activity. Contrary to expectation, high-affinity (13)NH(4)(+) influx in the amt1;1:T-DNA mutant was similar to the wild type when grown with adequate N. Removal of N to increase AtAMT1;1 expression decreased high-affinity (13)NH(4)(+) influx in the mutant by 30% compared with wild-type plants, whereas low-affinity (13)NH(4)(+) influx (250 microM-10 mM NH(4)(+)) exceeded that of wild-type plants. In these N-deprived plants, mRNA copy numbers of root AtAMT1;3 and AtAMT2;1 mRNA were significantly more increased in the mutant than in wild-type plants. Under most growth conditions, amt1;1:T-DNA plants were indistinguishable from the wild type, however, leaf morphology was altered. However, when grown with NH(4)(+) and sucrose, the mutant grew poorly and died. Our results are the first in planta evidence that AtAMT1;1 is a root NH(4)(+) transporter and that redundancies within the AMT family may allow compensation for the loss of AtAMT1;1.
Figures





Similar articles
-
Reciprocal leaf and root expression of AtAmt1.1 and root architectural changes in response to nitrogen starvation.Plant Physiol. 2007 Jan;143(1):236-50. doi: 10.1104/pp.106.088500. Epub 2006 Nov 3. Plant Physiol. 2007. PMID: 17085512 Free PMC article.
-
AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels.Plant J. 1999 Jul;19(2):143-52. doi: 10.1046/j.1365-313x.1999.00505.x. Plant J. 1999. PMID: 10476061
-
Differential regulation of the NO3- and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant.Plant J. 2001 Apr;26(2):143-55. doi: 10.1046/j.1365-313x.2001.01016.x. Plant J. 2001. PMID: 11389756
-
The regulation of nitrate and ammonium transport systems in plants.J Exp Bot. 2002 Apr;53(370):855-64. doi: 10.1093/jexbot/53.370.855. J Exp Bot. 2002. PMID: 11912228 Review.
-
The molecular physiology of ammonium uptake and retrieval.Curr Opin Plant Biol. 2000 Jun;3(3):254-61. Curr Opin Plant Biol. 2000. PMID: 10837267 Review.
Cited by
-
Leaf Amino Acid Supply Affects Photosynthetic and Plant Nitrogen Use Efficiency under Nitrogen Stress.Plant Physiol. 2018 Sep;178(1):174-188. doi: 10.1104/pp.18.00597. Epub 2018 Aug 6. Plant Physiol. 2018. PMID: 30082496 Free PMC article.
-
AtAMT1;4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis.Plant Cell Physiol. 2009 Jan;50(1):13-25. doi: 10.1093/pcp/pcn186. Epub 2008 Dec 10. Plant Cell Physiol. 2009. PMID: 19073648 Free PMC article.
-
Understanding the differential nitrogen sensing mechanism in rice genotypes through expression analysis of high and low affinity ammonium transporter genes.Mol Biol Rep. 2012 Mar;39(3):2233-41. doi: 10.1007/s11033-011-0972-2. Epub 2011 Jun 16. Mol Biol Rep. 2012. PMID: 21678052
-
Variation for N Uptake System in Maize: Genotypic Response to N Supply.Front Plant Sci. 2015 Nov 9;6:936. doi: 10.3389/fpls.2015.00936. eCollection 2015. Front Plant Sci. 2015. PMID: 26617612 Free PMC article.
-
Reciprocal leaf and root expression of AtAmt1.1 and root architectural changes in response to nitrogen starvation.Plant Physiol. 2007 Jan;143(1):236-50. doi: 10.1104/pp.106.088500. Epub 2006 Nov 3. Plant Physiol. 2007. PMID: 17085512 Free PMC article.
References
-
- Campisi L, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. - PubMed
-
- Cataldo DA, Haroon M, Schrader LE, Youngs VL. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal. 1975;6:71–80.
-
- Cerezo M, Tillard P, Gojon A, Primo-Millo E, Garcia-Agustin P. Characterization and regulation of ammonium transport systems in citrus plants. Planta. 2001;214:97–105. - PubMed
-
- Cohen BA, Wiles R, Campbell C. Pouring It On: Nitrate Contamination of Drinking Water. Washington, DC: Environmental Working Group; 1996.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

