Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases
- PMID: 12428018
- PMCID: PMC166672
- DOI: 10.1104/pp.007146
Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases
Abstract
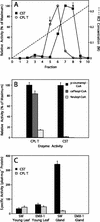
Sweet basil (Ocimum basilicum) peltate glandular trichomes produce a variety of small molecular weight phenylpropanoids, such as eugenol, caffeic acid, and rosmarinic acid, that result from meta hydroxylation reactions. Some basil lines do not synthesize eugenol but instead synthesize chavicol, a phenylpropanoid that does not contain a meta hydroxyl group. Two distinct acyltransferases, p-coumaroyl-coenzyme A:shikimic acid p-coumaroyl transferase and p-coumaroyl-coenzyme A:4-hydroxyphenyllactic acid p-coumaroyl transferase, responsible for the production of p-coumaroyl shikimate and of p-coumaroyl 4-hydroxyphenyllactate, respectively, were partially purified and shown to be specific for their substrates. p-Coumaroyl-coenzyme A:shikimic acid p-coumaroyl transferase is expressed in basil peltate glands that are actively producing eugenol and is not active in glands of noneugenol-producing basil plants, suggesting that the levels of this activity determine the levels of synthesis of some meta-hydroxylated phenylpropanoids in these glands such as eugenol. Two basil cDNAs encoding isozymes of cytochrome P450 CYP98A13, which meta hydroxylates p-coumaroyl shikimate, were isolated and found to be highly similar (90% identity) to the Arabidopsis homolog, CYP98A3. Like the Arabidopsis enzyme, the basil enzymes were found to be very specific for p-coumaroyl shikimate. Finally, additional hydroxylase activities were identified in basil peltate glands that convert p-coumaroyl 4-hydroxyphenyllactic acid to its caffeoyl derivative and p-coumaric acid to caffeic acid.
Figures




References
-
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. Cloning of three a-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moensch by a PCR approach and identification by expression in Escherichia coli of the CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol. 1998;36:393–405. - PubMed
-
- Beuerle T, Pichersky E. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem. 2002;302:305–312. - PubMed
-
- Chenchik A, Zhu Y, Diatchenko L, Li R, Hill J, Siebert P. Generation and use of high-quality cDNA from small amounts of total RNA by SMART PCR. In: Larrick J, editor. RT-PCR Methods for Gene Cloning and Analysis. Natick, MA: BioTechniques Books; 1996. pp. 305–319.
-
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002;30:33–45. - PubMed
-
- Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E. Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell. 2002;14:505–519. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

