The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism
- PMID: 12429060
- PMCID: PMC133445
- DOI: 10.1186/gb-2002-3-11-research0061
The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism
Abstract
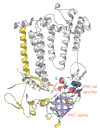
Background: The H subunit of the purple bacterial photosynthetic reaction center (PRC-H) is important for the assembly of the photosynthetic reaction center and appears to regulate electron transfer during the reduction of the secondary quinone. It contains a distinct cytoplasmic beta-barrel domain whose fold has no close structural relationship to any other well known beta-barrel domain.
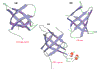
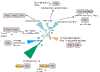
Results: We show that the PRC-H beta-barrel domain is the prototype of a novel superfamily of protein domains, the PRC-barrels, approximately 80 residues long, which is widely represented in bacteria, archaea and plants. This domain is also present at the carboxyl terminus of the pan-bacterial protein RimM, which is involved in ribosomal maturation and processing of 16S rRNA. A family of small proteins conserved in all known euryarchaea are composed entirely of a single stand-alone copy of the domain. Versions of this domain from photosynthetic proteobacteria contain a conserved acidic residue that is thought to regulate the reduction of quinones in the light-induced electron-transfer reaction. Closely related forms containing this acidic residue are also found in several non-photosynthetic bacteria, as well as in cyanobacteria, which have reaction centers with a different organization. We also show that the domain contains several determinants that could mediate specific protein-protein interactions.
Conclusions: The PRC-barrel is a widespread, ancient domain that appears to have been recruited to a variety of biological systems, ranging from RNA processing to photosynthesis. Identification of this versatile domain in numerous proteins could aid investigation of unexplored aspects of their biology.
Figures

References
-
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. - PubMed
-
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. - PubMed
-
- Orengo CA, Bray JE, Buchan DW, Harrison A, Lee D, Pearl FM, Sillitoe I, Todd AE, Thornton JM. The CATH protein family database: a resource for structural and functional annotation of genomes. Proteomics. 2002;2:11–21. - PubMed
-
- Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307:1113–1143. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

