Meiotic cohesion requires accumulation of ORD on chromosomes before condensation
- PMID: 12429833
- PMCID: PMC133601
- DOI: 10.1091/mbc.e02-06-0332
Meiotic cohesion requires accumulation of ORD on chromosomes before condensation
Erratum in
- Mol Biol Cell. 2003 Apr;14(4):following 1743
Abstract
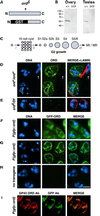
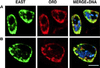
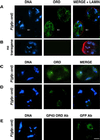
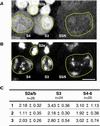
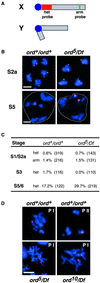
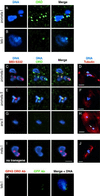
Cohesion between sister chromatids is a prerequisite for accurate chromosome segregation during mitosis and meiosis. To allow chromosome condensation during prophase, the connections that hold sister chromatids together must be maintained but still permit extensive chromatin compaction. In Drosophila, null mutations in the orientation disruptor (ord) gene lead to meiotic nondisjunction in males and females because cohesion is absent by the time that sister kinetochores make stable microtubule attachments. We provide evidence that ORD is concentrated within the extrachromosomal domains of the nuclei of Drosophila primary spermatocytes during early G2, but accumulates on the meiotic chromosomes by mid to late G2. Moreover, using fluorescence in situ hybridization to monitor cohesion directly, we show that cohesion defects first become detectable in ord(null) spermatocytes shortly after the time when wild-type ORD associates with the chromosomes. After condensation, ORD remains bound at the centromeres of wild-type spermatocytes and persists there until centromeric cohesion is released during anaphase II. Our results suggest that association of ORD with meiotic chromosomes during mid to late G2 is required to maintain sister-chromatid cohesion during prophase condensation and that retention of ORD at the centromeres after condensation ensures the maintenance of centromeric cohesion until anaphase II.
Figures
References
-
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. - PubMed
-
- Bickel SE, Orr-Weaver TL, Balicky EM. The sister-chromatid cohesion protein ORD is required for chiasma maintenance in Drosophila oocytes. Curr Biol. 2002;12:925–929. - PubMed
-
- Bickel SE, Orr-Weaver TL. Holding chromatids together to ensure they go their separate ways. BioEssays. 1996;18:293–300. - PubMed
-
- Bickel SE, Orr-Weaver TL. Regulation of sister-chromatid cohesion during Drosophila meiosis. In: Zirkin BR, editor. Germ Cell Development, Division, Disruption, and Death. New York: Springer-Verlag; 1998. pp. 37–48.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

