Microtubule flux and sliding in mitotic spindles of Drosophila embryos
- PMID: 12429839
- PMCID: PMC133607
- DOI: 10.1091/mbc.02-05-0069
Microtubule flux and sliding in mitotic spindles of Drosophila embryos
Abstract
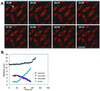
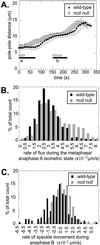
We proposed that spindle morphogenesis in Drosophila embryos involves progression through four transient isometric structures in which a constant spacing of the spindle poles is maintained by a balance of forces generated by multiple microtubule (MT) motors and that tipping this balance drives pole-pole separation. Here we used fluorescent speckle microscopy to evaluate the influence of MT dynamics on the isometric state that persists through metaphase and anaphase A and on pole-pole separation in anaphase B. During metaphase and anaphase A, fluorescent punctae on kinetochore and interpolar MTs flux toward the poles at 0.03 microm/s, too slow to drive chromatid-to-pole motion at 0.11 microm/s, and during anaphase B, fluorescent punctae on interpolar MTs move away from the spindle equator at the same rate as the poles, consistent with MT-MT sliding. Loss of Ncd, a candidate flux motor or brake, did not affect flux in the metaphase/anaphase A isometric state or MT sliding in anaphase B but decreased the duration of the isometric state. Our results suggest that, throughout this isometric state, an outward force exerted on the spindle poles by MT sliding motors is balanced by flux, and that suppression of flux could tip the balance of forces at the onset of anaphase B, allowing MT sliding and polymerization to push the poles apart.
Figures


References
-
- Endow SA, Chandra R, Komma DJ, Yamamoto AH, Salmon ED. Mutants of the Drosophila Ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J Cell Sci. 1994a;107:859–867. - PubMed
-
- Endow SA, Komma DJ. Centrosome and spindle function of the Drosophila Ncd motor protein visualized in live embryos using Ncd-GFP fusion proteins. J Cell Sci. 1996;109:2429–2442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

