The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization
- PMID: 12429848
- PMCID: PMC133616
- DOI: 10.1091/mbc.02-06-0092
The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization
Abstract
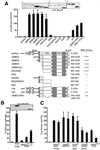
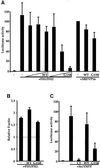
SRF-dependent transcription is regulated by the small GTPase RhoA via its effects on actin dynamics. The diaphanous-related formin (DRF) proteins have been identified as candidate RhoA effectors mediating signaling to SRF. Here we investigate the relationship between SRF activation and actin polymerization by the DRF mDia1. We show that the ability of mDia1 to potentiate SRF activity is strictly correlated with its ability to promote F-actin assembly. Both processes can occur independently of the mDia1 FH1 domain but require sequences in an extended C-terminal region encompassing the conserved FH2 domain. mDia-mediated SRF activation, but not F-actin assembly, can be blocked by a nonpolymerizable actin mutant, placing actin downstream of mDia in the signal pathway. The SRF activation assay was used to identify inactive mDia1 derivatives that inhibit serum- and LPA-induced signaling to SRF. We show that these interfering mutants also block F-actin assembly, whether induced by mDia proteins or extracellular signals. These results identify novel functional elements of mDia1 and show that it regulates SRF activity by inducing depletion of the cellular pool of G-actin.
Figures






References
-
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. - PubMed
-
- Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. - PubMed
-
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

