RAG-dependent peripheral T cell receptor diversification in CD8+ T lymphocytes
- PMID: 12432095
- PMCID: PMC137757
- DOI: 10.1073/pnas.242321099
RAG-dependent peripheral T cell receptor diversification in CD8+ T lymphocytes
Abstract
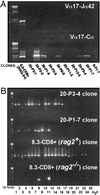
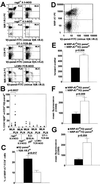
Rearrangement of T cell receptor (TCR) genes is driven by transient expression of V(D)J recombination-activating genes (RAGs) during lymphocyte development. Immunological dogma holds that T cells irreversibly terminate RAG expression before exiting the thymus, and that all of the progeny arising from mature T cells express the parental TCRs. When single pancreatic islet-derived, NRP-A7 peptide-reactive CD8(+) T cells from nonobese diabetic (NOD) mice were repeatedly stimulated with peptide-pulsed dendritic cells, daughter T cells reexpressed RAGs, lost their ability to bind to NRP-A7K(d) tetramers, ceased to transcribe tetramer-specific TCR genes, and, instead, expressed a vast array of other TCR rearrangements. Pancreatic lymph node (PLN) CD8(+) T cells from animals expressing a transgenic NRP-A7-reactive TCR transcribed and translated RAGs in vivo and displayed endogenous TCRs on their surface. RAG reexpression also occurred in the PLN CD8(+) T cells of wild-type NOD mice and could be induced in the peripheral CD8(+) T cells of nondiabetes-prone TCR-transgenic B10.H2(g7) mice by stimulation with peptide-pulsed dendritic cells. In contrast, reexpression of RAGs could not be induced in the CD8(+) T cells of B6 mice expressing an ovalbumin-specific, K(b)-restricted TCR, or in the CD8(+) T cells of NOD mice expressing a lymphocytic choriomeningitis virus-specific, D(b)-restricted TCR. Extra-thymic reexpression of the V(D)J recombination machinery in certain CD8(+) T cell subpopulations, therefore, enables further diversification of the peripheral T cell repertoire.
Figures

References
-
- Fugmann S., Lee, A., Shokett, P., Villey, I. & Schatz, D. (2000) Annu. Rev. Immunol. 18 495-527. - PubMed
-
- Nagaoka H., Yu, W. & Nussenzweig, M. (2000) Curr. Opin. Immunol. 12 187-190. - PubMed
-
- Turka L., Schatz, D., Oettinger, M., Chun, J., Gorka, C., Lee, K., McCormack, W. & Thompson, C. (1991) Science 16 778-781. - PubMed
-
- Borgulya P., Kishi, H., Uematsu, Y. & Boehmer, H. v. (1992) Cell 69 529-537. - PubMed
-
- Monroe R., Seidl, K., Gaertner, F., Han, S., Chen, F., Sekiguchi, J., Wang, J., Ferrini, R., Davidson, L., Kelsoe, G. & Alt, F. (1999) Immunity 11 201-212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

