Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae
- PMID: 12432101
- PMCID: PMC137751
- DOI: 10.1073/pnas.202604399
Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae
Abstract
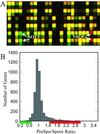
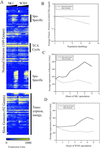
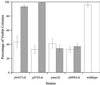
We have quantitatively monitored the sporulation and germination efficiencies of approximately 4,200 yeast deletion strains in parallel by using a molecular bar coding strategy. In a single study, we doubled the number of genes functionally implicated in sporulation to approximately 400, identifying both positive and negative regulators. Our set of 261 sporulation-deficient genes illustrates the importance of autophagy, carbon utilization, and transcriptional machinery during sporulation. These general cellular factors are more likely to exhibit fitness defects when deleted and less likely to be transcriptionally regulated than sporulation-specific genes. Our postgermination screening assay identified recombinationchromosome segregation genes, aneuploid strains, and possible germination-specific factors. Finally, our results facilitate a genome-wide comparison of expression pattern and mutant phenotype for a developmental process and suggest that 16% of genes differentially expressed during sporulation confer altered efficiency of spore production or defective postgermination growth when disrupted.
Figures

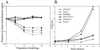
References
-
- Rabitsch K. P., Toth, A., Galova, M., Schleiffer, A., Schaffner, G., Aigner, E., Rupp, C., Penkner, A. M., Moreno-Borchart, A. C., Primig, M., et al. (2001) Curr. Biol. 11 1001-1009. - PubMed
-
- Briza P., Bogengruber, E., Thur, A., Rutzler, M., Munsterkotter, M., Dawes, I. W. & Breitenbach, M. (2002) Yeast 19 403-422. - PubMed
-
- Mitchell A. P. & Herskowitz, I. (1986) Nature 319 738-742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

