Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA
- PMID: 12433996
- PMCID: PMC137166
- DOI: 10.1093/nar/gkf618
Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA
Abstract
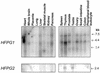
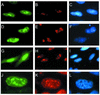
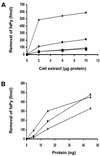
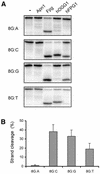
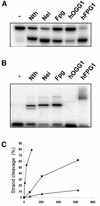
The mild phenotype associated with targeted disruption of the mouse OGG1 and NTH1 genes has been attributed to the existence of back-up activities and/or alternative pathways for the removal of oxidised DNA bases. We have characterised two new genes in human cells that encode DNA glycosylases, homologous to the bacterial Fpg (MutM)/Nei class of enzymes, capable of removing lesions that are substrates for both hOGG1 and hNTH1. One gene, designated HFPG1, showed ubiquitous expression in all tissues examined whereas the second gene, HFPG2, was only expressed at detectable levels in the thymus and testis. Transient transfections of HeLa cells with fusions of the cDNAs to EGFP revealed intracellular sorting to the nucleus with accumulation in the nucleoli for hFPG1, while hFPG2 co-localised with the 30 kDa subunit of RPA. hFPG1 was purified and shown to act on DNA substrates containing 8-oxoguanine, 5-hydroxycytosine and abasic sites. Removal of 8-oxoguanine, but not cleavage at abasic sites, was opposite base-dependent, with 8-oxoG:C being the preferred substrate and negligible activity towards 8-oxoG:A. It thus appears that hFPG1 has properties similar to mammalian OGG1 in preventing mutations arising from misincorporation of A across 8-oxoG and could function as a back-up repair activity for OGG1 in ogg1(-/-) mice.
Figures



References
-
- Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. - PubMed
-
- Kasai H., Crain,P.F., Kuchino,Y., Nishimura,S., Ootsuyama,A. and Tanooka,H. (1986) Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis, 7, 1849–1851. - PubMed
-
- Kasai H. and Nishimura,S. (1991) Formation of 8-hydroxydeoxyguanosine in DNA by oxygen radicals and its biological significance. In Sies,H. (ed.), Oxidative Stress: Oxidants and Antioxidants. Academic Press, London, UK, pp. 99–116.
-
- Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxoG. Nature, 349, 431–434. - PubMed
-
- Maki H. and Sekiguchi,M. (1992) MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature, 355, 273–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

