Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid
- PMID: 12434002
- PMCID: PMC137176
- DOI: 10.1093/nar/gkf630
Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid
Abstract
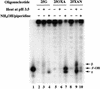
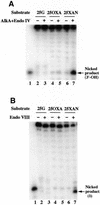
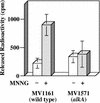
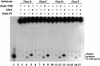
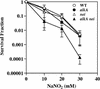
Nitrosation of guanine in DNA by nitrogen oxides such as nitric oxide (NO) and nitrous acid leads to formation of xanthine (Xan) and oxanine (Oxa), potentially cytotoxic and mutagenic lesions. In the present study, we have examined the repair capacity of DNA N-glycosylases from Escherichia coli for Xan and Oxa. The nicking assay with the defined substrates containing Xan and Oxa revealed that AlkA [in combination with endonuclease (Endo) IV] and Endo VIII recognized Xan in the tested enzymes. The activity (V(max)/K(m)) of AlkA for Xan was 5-fold lower than that for 7-methylguanine, and that of Endo VIII was 50-fold lower than that for thymine glycol. The activity of AlkA and Endo VIII for Xan was further substantiated by the release of [(3)H]Xan from the substrate. The treatment of E.coli with N-methyl-N'-nitro-N-nitrosoguanidine increased the Xan-excising activity in the cell extract from alkA(+) but not alkA(-) strains. The alkA and nei (the Endo VIII gene) double mutant, but not the single mutants, exhibited increased sensitivity to nitrous acid relative to the wild type strain. AlkA and Endo VIII also exhibited excision activity for Oxa, but the activity was much lower than that for Xan.
Figures
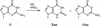


References
-
- Frederico L.A., Kunkel,T.A. and Shaw,B.R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry, 29, 2532–2537. - PubMed
-
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. - PubMed
-
- Hill-Perkins M., Jones,M.D. and Karran,P. (1986) Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat. Res., 162, 153–163. - PubMed
-
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

