Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia
- PMID: 12434020
- PMCID: PMC137750
- DOI: 10.1073/pnas.242606799
Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia
Abstract
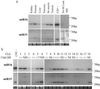
Micro-RNAs (miR genes) are a large family of highly conserved noncoding genes thought to be involved in temporal and tissue-specific gene regulation. MiRs are transcribed as short hairpin precursors ( approximately 70 nt) and are processed into active 21- to 22-nt RNAs by Dicer, a ribonuclease that recognizes target mRNAs via base-pairing interactions. Here we show that miR15 and miR16 are located at chromosome 13q14, a region deleted in more than half of B cell chronic lymphocytic leukemias (B-CLL). Detailed deletion and expression analysis shows that miR15 and miR16 are located within a 30-kb region of loss in CLL, and that both genes are deleted or down-regulated in the majority ( approximately 68%) of CLL cases.
Figures


References
-
- Dohner H., Stilgenbauer, S., Benner, A., Leupolt, E., Krober, A., Bullinger, L., Dohner, K., Bentz, M. & Lichter, P. (2000) N. Engl. J. Med. 343 1910-1916. - PubMed
-
- Bullrich F. & Croce, C. M. (2001) in Chronic Lymphoid Leukemias, ed. Chenson, B. D. (Dekker, New York), pp. 9–32.
-
- Dong J. T., Boyd, J. C. & Frierson, H. F., Jr. (2001) Prostate 49 166-171. - PubMed
-
- Bullrich F., Fujii, H., Calin, G., Mabuchi, H., Negrini, M., Pekarsky, Y., Rassenti, L., Alder, H., Reed, J. C., Keating, M. J., et al. (2001) Cancer Res. 61 6640-6648. - PubMed
-
- Migliazza A., Bosch, F., Komatsu, H., Cayanis, E., Martinotti, S., Toniato, E., Guccione, E., Qu, X., Chien, M., Murty, V. V., et al. (2001) Blood 97 2098-2104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

