REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export
- PMID: 12438415
- PMCID: PMC2173090
- DOI: 10.1083/jcb.200207128
REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export
Abstract
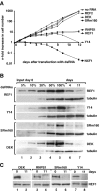
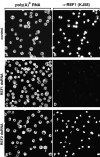
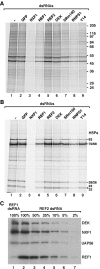
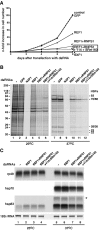
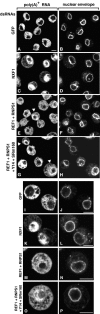
The metazoan proteins UAP56, REF1, and NXF1 are thought to bind sequentially to mRNA to promote its export to the cytoplasm: UAP56 is thought to recruit REF1 to nascent mRNA; REF1 acts as an adaptor protein mediating the association of NXF1 with mRNA, whereas NXF1 translocates the mRNA across the nuclear pore complex. REF1 is a component of the exon-exon junction complex (EJC); thus, the EJC is thought to play a role in the export of spliced mRNA. NXF1 and UAP56 are essential for mRNA export. An essential role for metazoan REF1 or the additional EJC proteins in this process has not been established. Contrary to expectation, we show that REF1 and the additional components of the EJC are dispensable for export of bulk mRNA in Drosophila cells. Only when REF1 and RNPS1 are codepleted, or when all EJC proteins are simultaneously depleted is a partial nuclear accumulation of polyadenylated RNAs observed. Because a significant fraction of bulk mRNA is detected in the cytoplasm of cells depleted of all EJC proteins, we conclude that additional adaptor protein(s) mediate the interaction between NXF1 and cellular mRNAs in metazoa. Our results imply that the essential role of UAP56 in mRNA export is not restricted to the recruitment of REF1.
Figures

References
-
- Adams, M.D., S.E. Celniker, R.A. Holt, C.A. Evans, J.D. Gocayne, P.G. Amanatides, S.E. Scherer, P.W. Li, R.A. Hoskins, et al. 2000. The genome sequence of Drosophila melanogaster. Science. 287:2185–2195. - PubMed
-
- Aravind, L., and E.V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

