Silencing of B cell receptor signals in human naive B cells
- PMID: 12438421
- PMCID: PMC2193982
- DOI: 10.1084/jem.20020881
Silencing of B cell receptor signals in human naive B cells
Retraction in
-
Retraction.J Exp Med. 2006 Nov 27;203(12):2779. doi: 10.1084/jem.2002088173106c. Epub 2006 Oct 30. J Exp Med. 2006. PMID: 17074927 Free PMC article. No abstract available.
Abstract
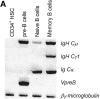
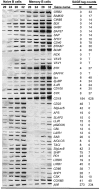
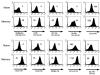
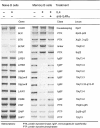
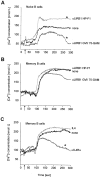
To identify changes in the regulation of B cell receptor (BCR) signals during the development of human B cells, we generated genome-wide gene expression profiles using the serial analysis of gene expression (SAGE) technique for CD34(+) hematopoietic stem cells (HSCs), pre-B cells, naive, germinal center (GC), and memory B cells. Comparing these SAGE profiles, genes encoding positive regulators of BCR signaling were expressed at consistently lower levels in naive B cells than in all other B cell subsets. Conversely, a large group of inhibitory signaling molecules, mostly belonging to the immunoglobulin superfamily (IgSF), were specifically or predominantly expressed in naive B cells. The quantitative differences observed by SAGE were corroborated by semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) and flow cytometry. In a functional assay, we show that down-regulation of inhibitory IgSF receptors and increased responsiveness to BCR stimulation in memory as compared with naive B cells at least partly results from interleukin (IL)-4 receptor signaling. Conversely, activation or impairment of the inhibitory IgSF receptor LIRB1 affected BCR-dependent Ca(2+) mobilization only in naive but not memory B cells. Thus, LIRB1 and IL-4 may represent components of two nonoverlapping gene expression programs in naive and memory B cells, respectively: in naive B cells, a large group of inhibitory IgSF receptors can elevate the BCR signaling threshold to prevent these cells from premature activation and clonal expansion before GC-dependent affinity maturation. In memory B cells, facilitated responsiveness upon reencounter of the immunizing antigen may result from amplification of BCR signals at virtually all levels of signal transduction.
Figures





References
-
- Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. - PubMed
-
- Lam, K.P., R. Kühn, and K. Rajewsky. 1997. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 90:1073–1083. - PubMed
-
- Kurosaki, T. 1999. Genetic analysis of B cell antigen receptor signaling. Annu. Rev. Immunol. 17:555–592. - PubMed
-
- Tarakhovsky, A., M. Turner, S. Schaal, P.J. Mee, L.P. Duddy, K. Rajewsky, and V.L. Tybulewicz. 1995. Defective antigen receptor-mediated proliferation of B and T cells in the absence of VAV. Nature. 374:467–470. - PubMed
-
- Ravetch, J.V., and L.L. Lanier. 2000. Immune inhibitory receptors. Science. 290:84–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

