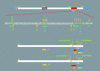
Deciphering the genesis and fate of amyloid beta-protein yields novel therapies for Alzheimer disease
- PMID: 12438432
- PMCID: PMC151820
- DOI: 10.1172/JCI16783
Deciphering the genesis and fate of amyloid beta-protein yields novel therapies for Alzheimer disease
Figures
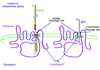
References
-
- Goedert M, Spillantini MG. Tau mutations in frontotemporal dementia FTDP-17 and their relevance for Alzheimer’s disease. Biochim Biophys Acta. 2000; 1502:110–121. - PubMed
-
- Selkoe DJ. Alzheimer’s disease: genes, proteins and therapies. Physiol Rev. 2001;81:742–761. - PubMed
-
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. - PubMed
-
- Buxbaum JD, et al. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem. 1998;273:27765–27767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

