Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization
- PMID: 12438575
- PMCID: PMC136672
- DOI: 10.1128/jvi.76.24.12491-12502.2002
Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization
Abstract
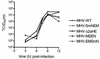
Despite their high frequency of RNA recombination, the plus-strand coronaviruses have a characteristic, strictly conserved genome organization with the essential genes occurring in the order 5'-polymerase (pol)-S-E-M-N-3'. We have investigated the significance of this remarkable conservation by rearrangement of the murine coronavirus genome through targeted recombination. Thus, viruses were prepared with the following gene order: 5'-pol-S-M-E-N-3', 5'-pol-S-N-E-M-3', 5'-pol-M-S-E-N-3', and 5'-pol-E-M-S-N-3'. All of these viruses were surprisingly viable, and most viruses replicated in cell culture with growth characteristics similar to those of the parental virus. The recombinant virus with the gene order 5'-pol-E-M-S-N-3' was also tested for the ability to replicate in the natural host, the mouse. The results indicate that the canonical coronavirus genome organization is not essential for replication in vitro and in vivo. Deliberate rearrangement of the viral genes may be useful in the generation of attenuated coronaviruses, which due to their reduced risk of generating viable viruses by recombination with circulating field viruses, would make safer vaccines.
Figures





References
-
- de Vries, A. A. F., A. L. Glaser, M. J. Raamsman, C. A. M. de Haan, S. Sarnataro, G. J. Godeke, and P. J. M. Rottier. 2000. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology 270:84-97. - PubMed
-
- de Vries, A. A. F., A. L. Glaser, M. J. Raamsman, and P. J. M. Rottier. 2001. Recombinant equine arteritis virus as an expression vector. Virology 284:259-276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

