The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription
- PMID: 12438606
- PMCID: PMC136662
- DOI: 10.1128/jvi.76.24.12813-12822.2002
The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription
Abstract
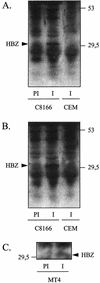
The RNA genome of the human T-cell leukemia virus type 1 (HTLV-1) codes for proteins involved in infectivity, replication, and transformation. We report in this study the characterization of a novel viral protein encoded by the complementary strand of the HTLV-1 RNA genome. This protein, designated HBZ (for HTLV-1 bZIP factor), contains a N-terminal transcriptional activation domain and a leucine zipper motif in its C terminus. We show here that HBZ is able to interact with the bZIP transcription factor CREB-2 (also called ATF-4), known to activate the HTLV-1 transcription by recruiting the viral trans-activator Tax on the Tax-responsive elements (TxREs). However, we demonstrate that the HBZ/CREB-2 heterodimers are no more able to bind to the TxRE and cyclic AMP response element sites. Taking these findings together, the functional inactivation of CREB-2 by HBZ is suggested to contribute to regulation of the HTLV-1 transcription. Moreover, the characterization of a minus-strand gene protein encoded by HTLV-1 has never been reported until now.
Figures






References
-
- Cibelli, G., S. Schoch, and G. Thiel. 1999. Nuclear targeting of cAMP response element binding protein 2 (CREB2). Eur. J. Cell Biol. 78:642-649. - PubMed
-
- Coudronniere, N., J. Corbeil, V. Robert-Hebmann, J. M. Mesnard, and C. Devaux. 1998. The lck protein tyrosine kinase is not involved in antibody-mediated CD4 (CDR3-loop) signal transduction that inhibits HIV-1 transcription. Eur. J. Immunol. 28:1445-1457. - PubMed
-
- Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225-21231. - PubMed
-
- Franklin, A. A., and J. K. Nyborg. 1995. Mechanisms of Tax regulation of human T cell leukemia virus type I gene expression. J. Biomed. Sci. 2:17-29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

