Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication
- PMID: 12438626
- PMCID: PMC136668
- DOI: 10.1128/jvi.76.24.13001-13014.2002
Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication
Abstract
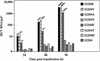
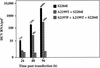
Hepatitis C virus (HCV) replication appears to be restricted to the human hepatoma cell line Huh-7, indicating that a favorable cellular environment exists within these cells. Although adaptive mutations in the HCV nonstructural proteins typically enhance the replicative capacity of subgenomic replicons in Huh-7 cells, replication can only be detected in a subpopulation of these cells. Here we show that self-replicating subgenomic RNA could be eliminated from Huh-7 clones by prolonged treatment with alpha interferon (IFN-alpha) and that a higher frequency of cured cells could support both subgenomic and full-length HCV replication. The increased permissiveness of one of the cured cell lines allowed us to readily detect HCV RNA and antigens early after RNA transfection, eliminating the need for selection of replication-positive cells. We also demonstrate that a single amino acid substitution in NS5A is sufficient for establishing HCV replication in a majority of cured cells and that the major phosphate acceptor site of subtype 1b NS5A is not essential for HCV replication.
Figures







References
-
- Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence and sequelae in hepatitis C virus infection: a perspective on the long-term outcome. Semin. Liver Dis. 20:17-25. - PubMed
-
- Bachmair, A., D. Finley, and A. Varshavasky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179-186. - PubMed
-
- Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. - PubMed
-
- Blight, K. J., A. Grakoui, H. L. Hanson, and C. M. Rice. 2002. The molecular biology of hepatitis C virus, p. 81-108. In J.-H. J. Ou (ed.), Hepatitis viruses. Kluwer Academic Publishers, Boston, Mass.
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

