Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism
- PMID: 12438647
- PMCID: PMC137721
- DOI: 10.1073/pnas.232517099
Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism
Erratum in
- Proc Natl Acad Sci U S A. 2003 Jan 21;100(2):763.
Abstract
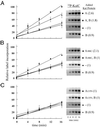
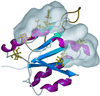
In the cyanobacterium Synechococcus elongatus (PCC 7942) the proteins KaiA, KaiB, and KaiC are required for circadian clock function. We deduced a circadian clock function for KaiA from a combination of biochemical and structural data. Both KaiA and its isolated carboxyl-terminal domain (KaiA180C) stimulated KaiC autophosphorylation and facilitated attenuation of KaiC autophosphorylation by KaiB. An amino-terminal domain (KaiA135N) had no function in the autophosphorylation assay. NMR structure determination showed that KaiA135N is a pseudo-receiver domain. We propose that this pseudo-receiver is a timing input-device that regulates KaiA stimulation of KaiC autophosphorylation, which in turn is essential for circadian timekeeping.
Figures



References
-
- Golden S. S., Johnson, C. H. & Kondo, T. (1998) Curr. Opin. Microbiol. 1 669-673. - PubMed
-
- Ishiura M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H. & Kondo, T. (1998) Science 281 1519-1523. - PubMed
-
- Young M. W. & Kay, S. A. (2001) Nat. Rev. Genet. 2 702-715. - PubMed
-
- Lorne J., Scheffer, J., Lee, A., Painter, M. & Miao, V. P. (2000) FEMS Microbiol. Lett. 189 129-133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

