Identification of the gene responsible for the cblA complementation group of vitamin B12-responsive methylmalonic acidemia based on analysis of prokaryotic gene arrangements
- PMID: 12438653
- PMCID: PMC137755
- DOI: 10.1073/pnas.242614799
Identification of the gene responsible for the cblA complementation group of vitamin B12-responsive methylmalonic acidemia based on analysis of prokaryotic gene arrangements
Abstract
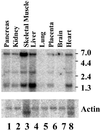
Vitamin B(12) (cobalamin) is an essential cofactor of two enzymes, methionine synthase and methylmalonyl-CoA mutase. The conversion of the vitamin to its coenzymes requires a series of biochemical modifications for which several genetic diseases are known, comprising eight complementation groups (cblA through cblH). The objective of this study was to clone the gene responsible for the cblA complementation group thought to represent a mitochondrial cobalamin reductase. Examination of bacterial operons containing genes in close proximity to the gene for methylmalonyl-CoA mutase and searching for orthologous sequences in the human genome yielded potential candidates. A candidate gene was evaluated for deleterious mutations in cblA patient cell lines, which revealed a 4-bp deletion in three cell lines, as well as an 8-bp insertion and point mutations causing a stop codon and an amino acid substitution. These data confirm that the identified gene, MMAA, corresponds to the cblA complementation group. It is located on chromosome 4q31.1-2 and encodes a predicted protein of 418 aa. A Northern blot revealed RNA species of 1.4, 2.6, and 5.5 kb predominating in liver and skeletal muscle. The deduced amino acid sequence reveals a domain structure, which belongs to the AAA ATPase superfamily that encompasses a wide variety of proteins including ATP-binding cassette transporter accessory proteins that bind ATP and GTP. We speculate that we have identified a component of a transporter or an accessory protein that is involved in the translocation of vitamin B(12) into mitochondria.
Figures






References
-
- Rosenblatt D. S. & Fenton, W. A. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Valle, D. & Sly, W. S. (McGraw–Hill, New York), pp. 3897–3933.
-
- Leclerc D., Odievre, M., Wu, Q., Wilson, A., Huizenga, J. J., Rozen, R., Scherer, S. W. & Gravel, R. A. (1999) Gene 240 75-88. - PubMed
-
- Leclerc D., Campeau, E., Goyette, P., Adjalla, C. E., Christensen, B., Ross, M., Eydoux, P., Rosenblatt, D. S., Rozen, R. & Gravel, R. A. (1996) Hum. Mol. Genet. 5 1867-1874. - PubMed
-
- Li Y. N., Gulati, S., Baker, P. J., Brody, L. C., Banerjee, R. & Kruger, W. D. (1996) Hum. Mol. Genet. 5 1851-1858. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

