A genomic-scale view of the cAMP response element-enhancer decoy: a tumor target-based genetic tool
- PMID: 12438686
- PMCID: PMC137767
- DOI: 10.1073/pnas.242617799
A genomic-scale view of the cAMP response element-enhancer decoy: a tumor target-based genetic tool
Abstract
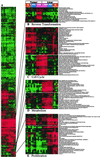
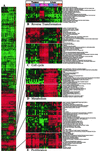
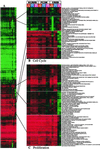
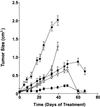
Enhancer DNA decoy oligodeoxynucleotides (ODNs) inhibit transcription by competing for transcription factors. A decoy ODN composed of the cAMP response element (CRE) inhibits CRE-directed gene transcription and tumor growth without affecting normal cell growth. Here, we use DNA microarrays to analyze the global effects of the CRE-decoy ODN in cancer cell lines and in tumors grown in nude mice. The CRE-decoy up-regulates the AP-2beta transcription factor gene in tumors but not in the livers of host animals. The up-regulated expression of AP-2beta is clustered with the up-regulation of other genes involved in development and cell differentiation. Concomitantly, another cluster of genes involved in cell proliferation and transformation is down-regulated. The observed alterations indicate that CRE-directed transcription favors tumor growth. The CRE-decoy ODN, therefore, may serve as a target-based genetic tool to treat cancer and other diseases in which CRE-directed transcription is abnormally used.
Figures
References
-
- Roesler W. J., Vandenbark, G. R. & Hanson, R. W. (1988) J. Biol. Chem. 263 9063-9066. - PubMed
-
- Park Y. G., Nesterova, M., Agrawal, S. & Cho-Chung, Y. S. (1999) J. Biol. Chem. 274 1573-1580. - PubMed
-
- Montminy M. R. & Bilezikjian, L. M. (1987) Nature 328 175-178. - PubMed
-
- Hai T. W., Liu, F., Allegretto, E. A., Karin, M. & Green, M. R. (1988) Genes Dev. 2 1216-1226. - PubMed
-
- Chrivia J. C., Kwok, R. P., Lamb, N., Hagiwara, M., Montminy, M. R. & Goodman, R. H. (1993) Nature 365 855-859. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

