A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades
- PMID: 12438687
- PMCID: PMC137760
- DOI: 10.1073/pnas.242608099
A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades
Abstract
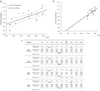
The prevalence of hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC) is considerably lower in the U.S. than in Japan. To elucidate this difference, we determined the time origin of the HCV epidemic in each country by using molecularly clocked long-term serial samples obtained from HCV carriers of genotypes 1a and 1b. The molecular clock estimated that HCV genotype 1 first appeared in Japan in around 1882, whereas emergence in the U.S. was delayed until around 1910. In addition, by statistical analysis using coalescent theory, the major spread time for HCV infection in Japan occurred in the 1930s, whereas widespread dissemination of HCV in the U.S. occurred in the 1960s. These estimates of viral spread time are consistent with epidemiologic observations and predict that the burden of HCC in the U.S. will increase in the next two to three decades, possibly to equal that currently experienced in Japan.
Figures



References
-
- Alter M. J. (1995) Semin. Liver Dis. 15 5-14. - PubMed
-
- Mansell C. J. & Locarnini, S. A. (1995) Semin. Liver Dis. 15 15-32. - PubMed
-
- Nishioka K., Watanabe, J., Furuta, S., Tanaka, E., Iino, S., Suzuki, H., Tsuji, T., Yano, M., Kuo, G., Choo, Q. L., et al. (1991) Cancer 67 429-433. - PubMed
-
- Shiratori Y., Shiina, S., Imamura, M., Kato, N., Kanai, F., Okudaira, T., Teratani, T., Tohgo, G., Toda, N., Ohashi, M., et al. (1995) Hepatology 22 1027-1033. - PubMed
-
- Kiyosawa K., Sodeyama, T., Tanaka, E., Gibo, Y., Yoshizawa, K., Nakano, Y., Furuta, S., Akahane, Y., Nishioka, K., Purcell, R. H., et al. (1990) Hepatology 12 671-675. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

