Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish
- PMID: 12438694
- PMCID: PMC137738
- DOI: 10.1073/pnas.232444199
Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish
Abstract
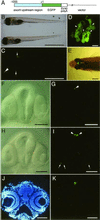
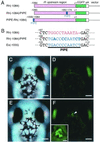
The pineal gland, sharing morphological and biochemical similarities with the retina, plays a unique and central role in the photoneuroendocrine system. The unique development of the pineal gland is directed by a specific combination of the expressed genes, but little is known about the regulatory mechanism underlying the pineal-specific gene expression. We isolated a 1.1-kbp fragment upstream of the zebrafish exo-rhodopsin (exorh) gene, which is expressed specifically in the pineal gland. Transgenic analysis using an enhanced green fluorescent protein reporter gene demonstrated that the proximal 147-bp region of the exorh promoter is sufficient to direct pineal-specific expression. This region contains three copies of a putative cone rod homeobox (Crx)Otx-binding site, which is known to be required for expression of both retina- and pineal-specific genes. Deletion and mutational analyses of the exorh promoter revealed that a previously uncharacterized sequence TGACCCCAATCT termed pineal expression-promoting element (PIPE) is required for pineal-specific promoter activity in addition to the CrxOtx-binding sites. By using the zebrafish rhodopsin (rh) promoter that drives retina-specific expression, we created a reporter construct having ectopic PIPE in the rh promoter at a position equivalent to that in the exorh promoter by introducing five nucleotide changes. Such a slight modification in the rh promoter induced ectopic enhanced green fluorescent protein expression in the pineal gland without affecting its retinal expression. These results identify PIPE as a critical cis-element contributing to the pineal-specific gene expression, in combination with the CrxOtx-binding site(s).
Figures

References
-
- Pu G. A. & Dowling, J. E. (1981) J. Neurophysiol. 46 1018-1038. - PubMed
-
- Cole W. C. & Youson, J. H. (1982) Am. J. Anat. 165 131-163. - PubMed
-
- Tamotsu S. & Morita, Y. (1986) J. Comp. Physiol. A 159 1-5. - PubMed
-
- Collin J. P., Voisin, P., Falcón, J., Faure, J. P., Brisson, P. & Defaye, J. R. (1989) Arch. Histol. Cytol. 52 441-449. - PubMed
-
- Shedpure M. & Pati, A. K. (1995) Indian J. Exp. Biol. 33 625-640. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

