Genomic analysis of the host response to hepatitis C virus infection
- PMID: 12441396
- PMCID: PMC137774
- DOI: 10.1073/pnas.202608199
Genomic analysis of the host response to hepatitis C virus infection
Abstract
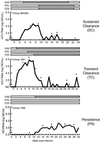
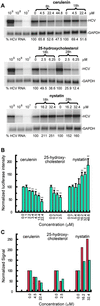
We have examined the progression of hepatitis C virus (HCV) infections by gene expression analysis of liver biopsies in acutely infected chimpanzees that developed persistent infection, transient viral clearance, or sustained clearance. Both common responses and outcome-specific changes in expression were observed. All chimpanzees showed gene expression patterns consistent with an IFN-alpha response that correlated with the magnitude and duration of infection. Transient and sustained viral clearance were uniquely associated with induction of IFN-gamma-induced genes and other genes involved in antigen processing and presentation and the adaptive immune response. During the early stages of infection, host genes involved in lipid metabolism were also differentially regulated. We also show that drugs that affect these biosynthetic pathways can regulate HCV replication in HCV replicon systems. Our results reveal genome-wide transcriptional changes that reflect the establishment, spread, and control of infection, and they reveal potentially unique antiviral programs associated with clearance of HCV infection.
Figures

References
-
- Hagedorn C. H. & Rice, C. M., (1999) Current Topics in Microbiology and Immunology (Springer, Berlin).
-
- Bartenschlager R. & Lohmann, V. (2001) Antiviral Res. 52 1-17. - PubMed
-
- Blight K. J., Kolykhalov, A. A. & Rice, C. M. (2000) Science 290 1972-1974. - PubMed
-
- Honda M., Kaneko, S., Kawai, H., Shirota, Y. & Kobayashi, K. (2001) Gastroenterology 120 955-966. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

