T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites
- PMID: 12446565
- PMCID: PMC1308325
- DOI: 10.1093/embo-reports/kvf237
T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites
Abstract
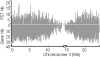
A statistical analysis of 9000 flanking sequence tags characterizing transferred DNA (T-DNA) transformants in Arabidopsis sheds new light on T-DNA insertion by illegitimate recombination. T-DNA integration is favoured in plant DNA regions with an A-T-rich content. The formation of a short DNA duplex between the host DNA and the left end of the T-DNA sets the frame for the recombination. The sequence immediately downstream of the plant A-T-rich region is the master element for setting up the DNA duplex, and deletions into the left end of the integrated T-DNA depend on the location of a complementary sequence on the T-DNA. Recombination at the right end of the T-DNA with the host DNA involves another DNA duplex, 2-3 base pairs long, that preferentially includes a G close to the right end of the T-DNA.
Figures


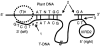
References
-
- Altschul S.F., Gish W., Miller W., Myers E.W. and Lipman D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
-
- Azpiroz-Leehan R. and Feldmann K.A. (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet., 13, 152–156. - PubMed
-
- Balzergue S. et al. (2001) Improved PCR-Walking for largescale isolation of plant T-DNA borders. Biotechniques, 30, 496–504. - PubMed
-
- Bechtold N., Ellis J. and Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by filtration of adult Arabidopsis thaliana plants. CR Acad. Sci. (Paris), 316, 1194–1199.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

