Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila
- PMID: 12446571
- PMCID: PMC1308319
- DOI: 10.1093/embo-reports/kvf243
Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila
Abstract
C2H2 zinc-finger proteins (ZFPs) constitute the largest family of nucleic acid binding factors in higher eukaryotes. In silico analysis identified a total of 326 putative ZFP genes in the Drosophila genome, corresponding to approximately 2.3% of the annotated genes. Approximately 29% of the Drosophila ZFPs are evolutionary conserved in humans and/or Caenorhabditis elegans. In addition, approximately 28% of the ZFPs contain an N-terminal zinc-finger-associated C4DM domain (ZAD) consisting of approximately 75 amino acid residues. The ZAD is restricted to ZFPs of dipteran and closely related insects. The evolutionary restriction, an expansion of ZAD-containing ZFP genes in the Drosophila genome and their clustering at few chromosomal sites are features reminiscent of vertebrate KRAB-ZFPs. ZADs are likely to represent protein-protein interaction domains. We propose that ZAD-containing ZFP genes participate in transcriptional regulation either directly or through site-specific modification and/or regulation of chromatin.
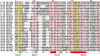
Figures
References
-
- Bhaskar V., Valentine S.A. and Courey A.J. (2000) A functional interaction between dorsal and components of the Smt3 conjugation machinery. J. Biol. Chem., 275, 4033–4040. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

