Mycobacterium bovis BCG response regulator essential for hypoxic dormancy
- PMID: 12446625
- PMCID: PMC135468
- DOI: 10.1128/JB.184.24.6760-6767.2002
Mycobacterium bovis BCG response regulator essential for hypoxic dormancy
Abstract
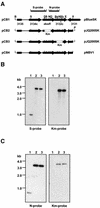
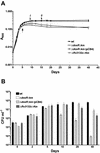
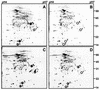
Obligately aerobic tubercle bacilli are capable of adapting to survive hypoxia by developing into a nonreplicating or dormant form. Dormant bacilli maintain viability for extended periods. Furthermore, they are resistant to antimycobacterials, and hence, dormancy might play a role in the persistence of tuberculosis infection despite prolonged chemotherapy. Previously, we have grown dormant Mycobacterium bovis BCG in an oxygen-limited Wayne culture system and subjected the bacilli to proteome analysis. This work revealed the upregulation of the response regulator Rv3133c and three other polypeptides (alpha-crystallin and two "conserved hypothetical" proteins) upon entry into dormancy. Here, we replaced the coding sequence of the response regulator with a kanamycin resistance cassette and demonstrated that the loss-of-function mutant died after oxygen starvation-induced termination of growth. Thus, the disruption of this dormancy-induced transcription factor resulted in loss of the ability of BCG to adapt to survival of hypoxia. Two-dimensional gel electrophoresis of protein extracts from the gene-disrupted strain showed that the genetic loss of the response regulator caused loss of the induction of the other three dormancy proteins. Thus, the upregulation of these dormancy proteins requires the response regulator. Based on these two functions, dormancy survival and regulation, we named the Rv3133c gene dosR for dormancy survival regulator. Our results provide conclusive evidence that DosR is a key regulator in the oxygen starvation-induced mycobacterial dormancy response.
Figures
References
-
- Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, S. Squares, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. - PubMed
-
- Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterisation of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tubercle Lung Dis. 80:141-159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

