Role for radA/sms in recombination intermediate processing in Escherichia coli
- PMID: 12446634
- PMCID: PMC135464
- DOI: 10.1128/JB.184.24.6836-6844.2002
Role for radA/sms in recombination intermediate processing in Escherichia coli
Abstract
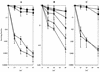
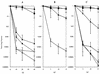
RadA/Sms is a highly conserved eubacterial protein that shares sequence similarity with both RecA strand transferase and Lon protease. We examined mutations in the radA/sms gene of Escherichia coli for effects on conjugational recombination and sensitivity to DNA-damaging agents, including UV irradiation, methyl methanesulfonate (MMS), mitomycin C, phleomycin, hydrogen peroxide, and hydroxyurea (HU). Null mutants of radA were modestly sensitive to the DNA-methylating agent MMS and to the DNA strand breakage agent phleomycin, with conjugational recombination decreased two- to threefold. We combined a radA mutation with other mutations in recombination genes, including recA, recB, recG, recJ, recQ, ruvA, and ruvC. A radA mutation was strongly synergistic with the recG Holliday junction helicase mutation, producing profound sensitivity to all DNA-damaging agents tested. Lesser synergy was noted between a mutation in radA and recJ, recQ, ruvA, ruvC, and recA for sensitivity to various genotoxins. For survival after peroxide and HU exposure, a radA mutation surprisingly suppressed the sensitivity of recA and recB mutants, suggesting that RadA may convert some forms of damage into lethal intermediates in the absence of these functions. Loss of radA enhanced the conjugational recombination deficiency conferred by mutations in Holliday junction-processing function genes, recG, ruvA, and ruvC. A radA recG ruv triple mutant had severe recombinational defects, to the low level exhibited by recA mutants. These results establish a role for RadA/Sms in recombination and recombinational repair, most likely involving the stabilization or processing of branched DNA molecules or blocked replication forks because of its genetic redundancy with RecG and RuvABC.
Figures



References
-
- Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd., vol. 2. ASM Press, Washington, D.C.
-
- Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science 232:485-487. - PubMed
-
- Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. - PubMed
-
- Byfield, J. E., Y. C. Lee, L. Tu, and F. Kulhanian. 1976. Molecular interactions of the combined effects of bleomycin and x-rays on mammalian cell survival. Cancer Res. 36:1138-1143. - PubMed
-
- Carrasco, B., S. Fernandez, K. Asai, N. Ogasawara, and C. Alonso. 2002. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genomics 266:899-906. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

