Iqg1p links spatial and secretion landmarks to polarity and cytokinesis
- PMID: 12446742
- PMCID: PMC2173104
- DOI: 10.1083/jcb.200205084
Iqg1p links spatial and secretion landmarks to polarity and cytokinesis
Abstract
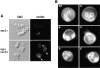
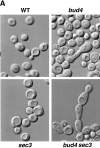
Cytokinesis requires the polarization of the actin cytoskeleton, the secretion machinery, and the correct positioning of the division axis. Budding yeast cells commit to their cytokinesis plane by choosing a bud site and polarizing their growth. Iqg1p (Cyk1p) was previously implicated in cytokinesis (Epp and Chant, 1997; Lippincott and Li, 1998; Osman and Cerione, 1998), as well as in the establishment of polarity and protein trafficking (Osman and Cerione, 1998). To better understand how Iqg1p influences these processes, we performed a two-hybrid screen and identified the spatial landmark Bud4p as a binding partner. Iqg1p can be coimmunoprecipitated with Bud4p, and Bud4p requires Iqg1p for its proper localization. Iqg1p also appears to specify axial bud-site selection and mediates the proper localization of the septin, Cdc12p, as well as binds and helps localize the secretion landmark, Sec3p. The double mutants iqg1Deltasec3Delta and bud4Deltasec3Delta display defects in polarity, budding pattern and cytokinesis, and electron microscopic studies reveal that these cells have aberrant septal deposition. Taken together, these findings suggest that Iqg1p recruits landmark proteins to form a targeting patch that coordinates axial budding with cytokinesis.
Figures








Similar articles
-
The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast.Mol Biol Cell. 1999 Feb;10(2):283-96. doi: 10.1091/mbc.10.2.283. Mol Biol Cell. 1999. PMID: 9950677 Free PMC article.
-
Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates cdc42p effects on the actin cytoskeleton.J Cell Biol. 1998 Jul 27;142(2):443-55. doi: 10.1083/jcb.142.2.443. J Cell Biol. 1998. PMID: 9679143 Free PMC article.
-
Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Oct;22(19):6906-20. doi: 10.1128/MCB.22.19.6906-6920.2002. Mol Cell Biol. 2002. PMID: 12215547 Free PMC article.
-
Regulation of septin organization and function in yeast.Trends Cell Biol. 2003 Aug;13(8):403-9. doi: 10.1016/s0962-8924(03)00151-x. Trends Cell Biol. 2003. PMID: 12888292 Review.
-
The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae.Cell Mol Life Sci. 2016 Aug;73(16):3115-36. doi: 10.1007/s00018-016-2220-3. Epub 2016 Apr 16. Cell Mol Life Sci. 2016. PMID: 27085703 Free PMC article. Review.
Cited by
-
A Flow Cytometry-Based Phenotypic Screen To Identify Novel Endocytic Factors in Saccharomyces cerevisiae.G3 (Bethesda). 2018 May 4;8(5):1497-1512. doi: 10.1534/g3.118.200102. G3 (Bethesda). 2018. PMID: 29540444 Free PMC article.
-
Spatial control of exocytosis.Curr Opin Cell Biol. 2003 Aug;15(4):430-7. doi: 10.1016/s0955-0674(03)00074-7. Curr Opin Cell Biol. 2003. PMID: 12892783 Free PMC article. Review.
-
A conserved role of IQGAP1 in regulating TOR complex 1.J Cell Sci. 2012 Apr 15;125(Pt 8):2041-52. doi: 10.1242/jcs.098947. Epub 2012 Feb 10. J Cell Sci. 2012. PMID: 22328503 Free PMC article.
-
Some assembly required: yeast septins provide the instruction manual.Trends Cell Biol. 2005 Aug;15(8):414-24. doi: 10.1016/j.tcb.2005.06.007. Trends Cell Biol. 2005. PMID: 16009555 Free PMC article. Review.
-
A molecular rheostat at the interface of cancer and diabetes.Biochim Biophys Acta. 2013 Aug;1836(1):166-76. doi: 10.1016/j.bbcan.2013.04.005. Epub 2013 Apr 29. Biochim Biophys Acta. 2013. PMID: 23639840 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

