A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3' processing and cell cycle regulation
- PMID: 12446762
- PMCID: PMC139887
- DOI: 10.1128/MCB.22.24.8415-8425.2002
A sequence element downstream of the yeast HTB1 gene contributes to mRNA 3' processing and cell cycle regulation
Erratum in
- Mol Cell Biol. 2008 Mar;28(5):1873
Abstract
Histone mRNAs accumulate in the S phase and are rapidly degraded as cells progress into the G(2) phase of the cell cycle. In Saccharomyces cerevisiae, fusion of the 3' untranslated region and downstream sequences of the yeast histone gene HTB1 to a neomycin phosphotransferase open reading frame is sufficient to confer cell cycle regulation on the resulting chimera gene (neo-HTB1). We have identified a sequence element, designated the distal downstream element (DDE), that influences both the 3'-end cleavage site selection and the cell cycle regulation of the neo-HTB1 mRNA. Mutations in the DDE, which is located approximately 110 nucleotides downstream of the HTB1 gene, lead to a delay in the accumulation of the neo-HTB1 mRNA in the S phase and a lack of mRNA turnover in the G(2) phase. The DDE is transcribed as part of the primary transcript and binds a protein factor(s). Maximum binding is observed in the S phase of the cell cycle, and mutations that affect the turnover of the HTB1 mRNA alter the binding activity. While located in the same general region, mutations that affect 3'-end cleavage site selection act independently from those that alter the cell cycle regulation.
Figures






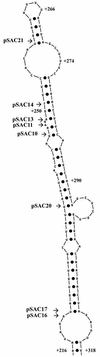
Similar articles
-
Coding and noncoding sequences at the 3' end of yeast histone H2B mRNA confer cell cycle regulation.Mol Cell Biol. 1990 Jun;10(6):2687-94. doi: 10.1128/mcb.10.6.2687-2694.1990. Mol Cell Biol. 1990. PMID: 2188095 Free PMC article.
-
Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae.Nucleic Acids Res. 2007;35(18):6268-79. doi: 10.1093/nar/gkm691. Epub 2007 Sep 13. Nucleic Acids Res. 2007. PMID: 17855393 Free PMC article.
-
Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast.Mol Cell Biol. 2011 Feb;31(3):557-72. doi: 10.1128/MCB.00909-10. Epub 2010 Nov 29. Mol Cell Biol. 2011. PMID: 21115727 Free PMC article.
-
CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis, and transcription during the cell cycle.J Biol Chem. 1992 Jun 5;267(16):11274-80. J Biol Chem. 1992. PMID: 1597462
-
Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae.Mol Cell Biol. 1993 Jan;13(1):28-38. doi: 10.1128/mcb.13.1.28-38.1993. Mol Cell Biol. 1993. PMID: 8417331 Free PMC article.
Cited by
-
Regulation of histone gene expression in budding yeast.Genetics. 2012 May;191(1):7-20. doi: 10.1534/genetics.112.140145. Genetics. 2012. PMID: 22555441 Free PMC article. Review.
-
The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres.Mol Cell Biol. 2004 May;24(10):4309-20. doi: 10.1128/MCB.24.10.4309-4320.2004. Mol Cell Biol. 2004. PMID: 15121850 Free PMC article.
-
Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae.Genetics. 2007 Mar;175(3):993-1010. doi: 10.1534/genetics.106.065987. Epub 2006 Dec 18. Genetics. 2007. PMID: 17179095 Free PMC article.
-
Recombination between homoeologous chromosomes of lager yeasts leads to loss of function of the hybrid GPH1 gene.Appl Environ Microbiol. 2009 Jul;75(13):4573-9. doi: 10.1128/AEM.00351-09. Epub 2009 May 8. Appl Environ Microbiol. 2009. PMID: 19429545 Free PMC article.
-
The PolyA tail length of yeast histone mRNAs varies during the cell cycle and is influenced by Sen1p and Rrp6p.Nucleic Acids Res. 2012 Mar;40(6):2700-11. doi: 10.1093/nar/gkr1108. Epub 2011 Nov 28. Nucleic Acids Res. 2012. PMID: 22123738 Free PMC article.
References
-
- Aranda, A., and N. Proudfoot. 2001. Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell 7:1003-1011. - PubMed
-
- Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, K. Keller, and N. Proudfoot. 1998. Coupling transcriptional termination to messenger RNA maturation in yeast. Science 280:298-301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
