Developmental changes in the Sciara II/9A initiation zone for DNA replication
- PMID: 12446763
- PMCID: PMC139883
- DOI: 10.1128/MCB.22.24.8426-8437.2002
Developmental changes in the Sciara II/9A initiation zone for DNA replication
Abstract
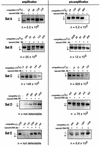
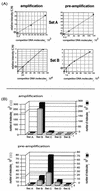
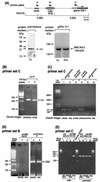
Developmentally regulated initiation of DNA synthesis was studied in the fly Sciara at locus II/9A. PCR analysis of nascent strands revealed an initiation zone that spans approximately 8 kb in mitotic embryonic cells and endoreplicating salivary glands but contracts to 1.2 to 2.0 kb during DNA amplification of DNA puff II/9A. Thus, the amplification origin occurs within the initiation zone used for normal replication. The initiation zone left-hand border is constant, but the right-hand border changes during development. Also, there is a shift in the preferred site for initiation of DNA synthesis during DNA amplification compared to that in preamplification stages. This is the first demonstration that once an initiation zone is defined in embryos, its borders and preferred replication start sites can change during development. Chromatin immunoprecipitation showed that the RNA polymerase II 140-kDa subunit occupies the promoter of gene II/9-1 during DNA amplification, even though intense transcription will not start until the next developmental stage. RNA polymerase II is adjacent to the right-hand border of the initiation zone at DNA amplification but not at preamplification, suggesting that it may influence the position of this border. These findings support a relationship between the transcriptional machinery and establishment of the replication initiation zone.
Figures



Similar articles
-
Initiation of DNA replication in multicellular eukaryotes.J Struct Biol. 2002 Oct-Dec;140(1-3):17-30. doi: 10.1016/s1047-8477(02)00538-5. J Struct Biol. 2002. PMID: 12490150 Review.
-
Ecdysone induces transcription and amplification in Sciara coprophila DNA puff II/9A.Dev Biol. 2006 Nov 1;299(1):151-63. doi: 10.1016/j.ydbio.2006.07.018. Epub 2006 Jul 21. Dev Biol. 2006. PMID: 16938289
-
DNA amplification in DNA puff II/9A of Sciara coprophila.Cold Spring Harb Symp Quant Biol. 1993;58:487-94. doi: 10.1101/sqb.1993.058.01.055. Cold Spring Harb Symp Quant Biol. 1993. PMID: 7956063 No abstract available.
-
Replication initiates at a confined region during DNA amplification in Sciara DNA puff II/9A.Genes Dev. 1993 Jun;7(6):1072-84. doi: 10.1101/gad.7.6.1072. Genes Dev. 1993. PMID: 8504930
-
DNA sequence amplification in sciarid flies: results and perspectives.Braz J Med Biol Res. 1991;24(3):233-48. Braz J Med Biol Res. 1991. PMID: 1823237 Review.
Cited by
-
The ecdysone receptor (ScEcR-A) binds DNA puffs at the start of DNA amplification in Sciara coprophila.Chromosome Res. 2013 Jul;21(4):345-60. doi: 10.1007/s10577-013-9360-1. Epub 2013 Jun 5. Chromosome Res. 2013. PMID: 23737076 Free PMC article.
-
Involvement of G-quadruplex regions in mammalian replication origin activity.Nat Commun. 2019 Jul 22;10(1):3274. doi: 10.1038/s41467-019-11104-0. Nat Commun. 2019. PMID: 31332171 Free PMC article.
-
The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries.Genes Dev. 2004 Feb 15;18(4):397-410. doi: 10.1101/gad.1171404. Epub 2004 Feb 20. Genes Dev. 2004. PMID: 14977920 Free PMC article.
-
In search of the holy replicator.Nat Rev Mol Cell Biol. 2004 Oct;5(10):848-55. doi: 10.1038/nrm1495. Nat Rev Mol Cell Biol. 2004. PMID: 15459665 Free PMC article. Review.
-
DNA replication: the unbearable lightness of origins.EMBO Rep. 2006 Aug;7(8):779-81. doi: 10.1038/sj.embor.7400766. EMBO Rep. 2006. PMID: 16880822 Free PMC article. Review. No abstract available.
References
-
- Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. - PubMed
-
- Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the α-globin locus control region in initiation of DNA replication. Science 270:815-819. - PubMed
-
- Aladjem, M. I., W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
