Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements
- PMID: 12446765
- PMCID: PMC139884
- DOI: 10.1128/MCB.22.24.8448-8456.2002
Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements
Abstract
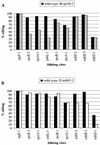
RNA editing in organelles of angiosperm plants results in alteration of Cs to Us in transcripts. In most editing sites analyzed in vitro or in vivo, sequences within approximately 30 nucleotides (nt) 5' and 10 nt 3' of the edited C have been found to be required for selection of the correct C editing target and for editing efficiency, but no consensus sequences have been identified. The effect of high-level expression of two different minigenes carrying either the rpoB-2 or the ndhF-2 editing site on editing was determined for all 31 known edited Cs in two transgenic tobacco lines. The editing efficiencies of both the corresponding rpoB and ndhF editing sites in the endogenous genes' transcripts and in several other genes' transcripts were reduced in the chloroplast transgenic plants. Conserved nucleotides could be identified in the sequences immediately 5' of each overexpressed editing site and in the sites in the additional genes that experienced a competition effect, though the conserved nucleotides differ 5' of rpoB-2 and 5' of ndhF-2. Inspection of sequences surrounding chloroplast and mitochondrial editing sites reveals that they can be grouped into clusters which carry conserved nucleotides within 30 nt 5' of the C target of editing. The data are consistent with a model in which the same trans factor recognizes several chloroplast or mitochondrial editing sites, depending on the particular sequence 5' of the edited C.
Figures






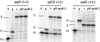
References
-
- Anant, S., and N. O. Davidson. 2000. An AU-rich sequence element (UUUN[A/U]U) downstream of the edited C in apolipoprotein B mRNA is a high-affinity binding site for Apobec-1: binding of Apobec-1 to this motif in the 3′ untranslated region of c-myc increases mRNA stability. Mol. Cell. Biol. 20:1982-1992. - PMC - PubMed
-
- Bock, R. 2002. RNA editing in plant mitochondria and chloroplasts, p. 38-60. In B. L. Bass (ed.), RNA editing. Oxford University Press, Oxford, England.
-
- Bock, R. 2000. Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82:549-557. - PubMed
-
- Bock, R., and P. Maliga. 1995. In vivo testing of a tobacco plastid DNA segment for guide RNA function in psbL editing. Mol. Gen. Genet. 247:439-443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
