Expression of a novel, sterol-insensitive form of sterol regulatory element binding protein 2 (SREBP2) in male germ cells suggests important cell- and stage-specific functions for SREBP targets during spermatogenesis
- PMID: 12446768
- PMCID: PMC139869
- DOI: 10.1128/MCB.22.24.8478-8490.2002
Expression of a novel, sterol-insensitive form of sterol regulatory element binding protein 2 (SREBP2) in male germ cells suggests important cell- and stage-specific functions for SREBP targets during spermatogenesis
Abstract
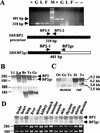
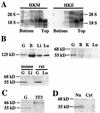
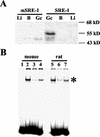
Cholesterol biosynthesis in somatic cells is controlled at the transcriptional level by a homeostatic feedback pathway involving sterol regulatory element binding proteins (SREBPs). These basic helix-loop-helix (bHLH)-Zip proteins are synthesized as membrane-bound precursors, which are cleaved to form a soluble, transcriptionally active mature SREBP that regulates the promoters for genes involved in lipid synthesis. Homeostasis is conferred by sterol feedback inhibition of this maturation process. Previous work has demonstrated the expression of SREBP target genes in the male germ line, several of which are highly up-regulated during specific developmental stages. However, the role of SREBPs in the control of sterol regulatory element-containing promoters during spermatogenesis has been unclear. In particular, expression of several of these genes in male germ cells appears to be insensitive to sterols, contrary to SREBP-dependent gene regulation in somatic cells. Here, we have characterized a novel isoform of the transcription factor SREBP2, which is highly enriched in rat and mouse spermatogenic cells. This protein, SREBP2gc, is expressed in a stage-dependent fashion as a soluble, constitutively active transcription factor that is not subject to feedback control by sterols. These findings likely explain the apparent sterol-insensitive expression of lipid synthesis genes during spermatogenesis. Expression of a sterol-independent, constitutively active SREBP2gc in the male germ line may have arisen as a means to regulate SREBP target genes in specific developmental stages. This may reflect unique roles for cholesterol synthesis and other functional targets of SREBPs during spermatogenesis.
Figures

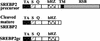


Similar articles
-
Novel role for a sterol response element binding protein in directing spermatogenic cell-specific gene expression.Mol Cell Biol. 2004 Dec;24(24):10681-8. doi: 10.1128/MCB.24.24.10681-10688.2004. Mol Cell Biol. 2004. PMID: 15572673 Free PMC article.
-
SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element.Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11603-7. doi: 10.1073/pnas.90.24.11603. Proc Natl Acad Sci U S A. 1993. PMID: 7903453 Free PMC article.
-
Lanosterol metabolism and sterol regulatory element binding protein (SREBP) expression in male germ cell maturation.J Steroid Biochem Mol Biol. 2003 Jun;85(2-5):429-38. doi: 10.1016/s0960-0760(03)00191-2. J Steroid Biochem Mol Biol. 2003. PMID: 12943732
-
SREBP transcription factors: master regulators of lipid homeostasis.Biochimie. 2004 Nov;86(11):839-48. doi: 10.1016/j.biochi.2004.09.018. Biochimie. 2004. PMID: 15589694 Review.
-
Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins.World J Gastroenterol. 2004 Nov 1;10(21):3081-7. doi: 10.3748/wjg.v10.i21.3081. World J Gastroenterol. 2004. PMID: 15457548 Free PMC article. Review.
Cited by
-
Identification of novel genes and pathways regulating SREBP transcriptional activity.PLoS One. 2009;4(4):e5197. doi: 10.1371/journal.pone.0005197. Epub 2009 Apr 21. PLoS One. 2009. PMID: 19381295 Free PMC article.
-
Insights from the single-cell level: lineage trajectory and somatic-germline interactions during spermatogenesis in dwarf surfclam Mulinia lateralis.BMC Genomics. 2025 Jan 24;26(1):69. doi: 10.1186/s12864-025-11266-w. BMC Genomics. 2025. PMID: 39856558 Free PMC article.
-
Dual functions of Insig proteins in cholesterol homeostasis.Lipids Health Dis. 2012 Dec 18;11:173. doi: 10.1186/1476-511X-11-173. Lipids Health Dis. 2012. PMID: 23249523 Free PMC article. Review.
-
The Administration of Chitosan-Tripolyphosphate-DNA Nanoparticles to Express Exogenous SREBP1a Enhances Conversion of Dietary Carbohydrates into Lipids in the Liver of Sparus aurata.Biomolecules. 2019 Jul 24;9(8):297. doi: 10.3390/biom9080297. Biomolecules. 2019. PMID: 31344838 Free PMC article.
-
Implications of adiponectin in linking metabolism to testicular function.Endocrine. 2014 May;46(1):16-28. doi: 10.1007/s12020-013-0102-0. Epub 2013 Nov 28. Endocrine. 2014. PMID: 24287788 Review.
References
-
- Bajpai, M., G. Gupta, S. K. Jain, and B. S. Setty. 1998. Lipid metabolising enzymes in isolated rat testicular germ cells and changes associated with meiosis. Andrologia 30:311-315. - PubMed
-
- Beckman, J. K., and J. G. Coniglio. 1979. A comparative study of the lipid composition of isolated rat Sertoli and germinal cells. Lipids 14:262-267. - PubMed
-
- Bellve, A. R., C. F. Millette, Y. M. Bhatnagar, and D. A. O'Brien. 1977. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J. Histochem. Cytochem. 25:480-494. - PubMed
-
- Blendy, J. A., K. H. Kaestner, G. F. Weinbauer, E. Nieschlag, and G. Schutz. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380:162-165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
