Interdependent interactions between TFIIB, TATA binding protein, and DNA
- PMID: 12446790
- PMCID: PMC139873
- DOI: 10.1128/MCB.22.24.8735-8743.2002
Interdependent interactions between TFIIB, TATA binding protein, and DNA
Abstract
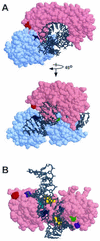
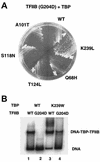
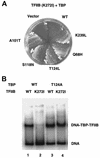
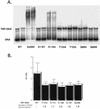
Temperature-sensitive mutants of TFIIB that are defective for essential interactions were isolated. One mutation (G204D) results in disruption of a protein-protein contact between TFIIB and TATA binding protein (TBP), while the other (K272I) disrupts an interaction between TFIIB and DNA. The TBP gene was mutagenized, and alleles that suppress the slow-growth phenotypes of the TFIIB mutants were isolated. TFIIB with the G204D mutation [TFIIB(G204D)] was suppressed by hydrophobic substitutions at lysine 239 of TBP. These changes led to increased affinity between TBP and TFIIB. TFIIB(K272I) was weakly suppressed by TBP mutants in which K239 was changed to hydrophobic residues. However, this mutant TFIIB was strongly suppressed by conservative substitutions in the DNA binding surface of TBP. Biochemical characterization showed that these TBP mutants had increased affinity for a TATA element. The TBPs with increased affinity could not suppress TFIIB(G204D), leading us to propose a two-step model for the interaction between TFIIB and the TBP-DNA complex.
Figures
Similar articles
-
Role of the inhibitory DNA-binding surface of human TATA-binding protein in recruitment of human TFIIB family members.Mol Cell Biol. 2003 Nov;23(22):8152-60. doi: 10.1128/MCB.23.22.8152-8160.2003. Mol Cell Biol. 2003. PMID: 14585974 Free PMC article.
-
Interactions of a DNA-bound transcriptional activator with the TBP-TFIIA-TFIIB-promoter quaternary complex.J Biol Chem. 2003 Mar 28;278(13):11495-501. doi: 10.1074/jbc.M211938200. Epub 2003 Jan 21. J Biol Chem. 2003. PMID: 12538582 Free PMC article.
-
Promoter-specific activation defects by a novel yeast TBP mutant compromised for TFIIB interaction.Curr Biol. 2001 Nov 13;11(22):1794-8. doi: 10.1016/s0960-9822(01)00566-8. Curr Biol. 2001. PMID: 11719223
-
X-ray crystallographic studies of eukaryotic transcription initiation factors.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):483-9. doi: 10.1098/rstb.1996.0046. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735270 Review.
-
Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation.Gene. 2022 Jul 30;833:146581. doi: 10.1016/j.gene.2022.146581. Epub 2022 May 18. Gene. 2022. PMID: 35597524 Review.
Cited by
-
TFIIB and the regulation of transcription by RNA polymerase II.Chromosoma. 2007 Oct;116(5):417-29. doi: 10.1007/s00412-007-0113-9. Epub 2007 Jun 26. Chromosoma. 2007. PMID: 17593382 Review.
-
Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3.Genes Dev. 2008 Nov 1;22(21):2994-3006. doi: 10.1101/gad.1724408. Genes Dev. 2008. PMID: 18981477 Free PMC article.
-
C-terminal hydrophobic interactions play a critical role in oligomeric assembly of the P22 tailspike trimer.Protein Sci. 2003 Dec;12(12):2732-47. doi: 10.1110/ps.03150303. Protein Sci. 2003. PMID: 14627734 Free PMC article.
-
Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8.Genetics. 2007 Dec;177(4):2007-17. doi: 10.1534/genetics.107.081976. Genetics. 2007. PMID: 18073420 Free PMC article.
-
The yeast FACT complex has a role in transcriptional initiation.Mol Cell Biol. 2005 Jul;25(14):5812-22. doi: 10.1128/MCB.25.14.5812-5822.2005. Mol Cell Biol. 2005. PMID: 15987999 Free PMC article.
References
-
- Ackers, G. K., M. A. Shea, and F. R. Smith. 1983. Free energy coupling within macromolecules. The chemical work of ligand binding at the individual sites in co-operative systems. J. Mol. Biol. 170:223-242. - PubMed
-
- Bagby, S., S. Kim, E. Maldonado, K. I. Tong, D. Reinberg, and M. Ikura. 1995. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell 82:857-867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
