The role of chromosomal instability in tumor initiation
- PMID: 12446840
- PMCID: PMC138593
- DOI: 10.1073/pnas.202617399
The role of chromosomal instability in tumor initiation
Abstract
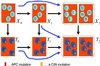
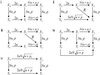
Chromosomal instability (CIN) is a defining characteristic of most human cancers. Mutation of CIN genes increases the probability that whole chromosomes or large fractions of chromosomes are gained or lost during cell division. The consequence of CIN is an imbalance in the number of chromosomes per cell (aneuploidy) and an enhanced rate of loss of heterozygosity. A major question of cancer genetics is to what extent CIN, or any genetic instability, is an early event and consequently a driving force for tumor progression. In this article, we develop a mathematical framework for studying the effect of CIN on the somatic evolution of cancer. Specifically, we calculate the conditions for CIN to initiate the process of colorectal tumorigenesis before the inactivation of tumor suppressor genes.
Figures

References
-
- Friend S. H., Bernards, R., Rogels, S., Weinberg, R. A., Rapaport, J. M., Albert, D. M. & Dryja, T. P. (1986) Nature 323, 643-646. - PubMed
-
- Lengauer C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643-649. - PubMed
-
- Sen S. (2000) Curr. Opin. Oncol. 12, 82-88. - PubMed
-
- Loeb L. A., Springgate, C. F. & Battula, N. (1974) Cancer Res. 34, 2311-2321. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

