Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti
- PMID: 12450806
- PMCID: PMC134414
- DOI: 10.1128/AEM.68.12.5877-5881.2002
Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti
Abstract
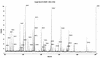
Rhizobia are soil bacteria that are able to establish symbiotic associations with leguminous hosts. In iron-limited environments these bacteria can use iron present in heme or heme compounds (hemoglobin, leghemoglobin). Here we report the presence in Sinorhizobium meliloti of an iron-regulated outer membrane protein that is able to bind hemin but not hemoglobin. Protein assignment was done by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Tryptic peptides correlated with the mass measurements obtained accounted for 54% of the translated sequence of a putative heme receptor gene present in the chromosome of S. meliloti 1021. The results which we obtained suggest that this protein (designated ShmR for Sinorhizobium heme receptor) is involved in high-affinity heme-mediated iron transport.
Figures
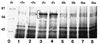


References
-
- Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dréano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thébault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. - PMC - PubMed
-
- Carson, K. C., A. R. Glenn, and M. J. Dilworth. 1994. Specificity of siderophore-mediated transport of iron in rhizobia. Arch. Microbiol. 161:333-339.
-
- Fabiano, E., G. Gualtieri, C. Pritch, G. Polla, and A. Arias. 1994. Extent of high-affinity iron transport systems in field isolates of rhizobia. Plant Soil 164:177-185.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

