Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds
- PMID: 12450818
- PMCID: PMC134431
- DOI: 10.1128/AEM.68.12.5973-5980.2002
Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds
Abstract
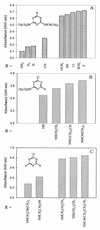
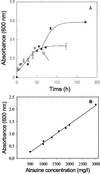
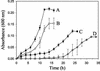
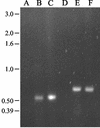
Arthrobacter aurescens strain TC1 was isolated without enrichment by plating atrazine-contaminated soil directly onto atrazine-clearing plates. A. aurescens TC1 grew in liquid medium with atrazine as the sole source of nitrogen, carbon, and energy, consuming up to 3,000 mg of atrazine per liter. A. aurescens TC1 is metabolically diverse and grew on a wider range of s-triazine compounds than any bacterium previously characterized. The 23 s-triazine substrates serving as the sole nitrogen source included the herbicides ametryn, atratone, cyanazine, prometryn, and simazine. Moreover, atrazine substrate analogs containing fluorine, mercaptan, and cyano groups in place of the chlorine substituent were also growth substrates. Analogs containing hydrogen, azido, and amino functionalities in place of chlorine were not growth substrates. A. aurescens TC1 also metabolized compounds containing chlorine plus N-ethyl, N-propyl, N-butyl, N-s-butyl, N-isobutyl, or N-t-butyl substituents on the s-triazine ring. Atrazine was metabolized to alkylamines and cyanuric acid, the latter accumulating stoichiometrically. Ethylamine and isopropylamine each served as the source of carbon and nitrogen for growth. PCR experiments identified genes with high sequence identity to atzB and atzC, but not to atzA, from Pseudomonas sp. strain ADP.
Figures


References
-
- Aspelin, A. L., and A. H. Grube. 1999. Pesticides industry sales and usage 1996 and 1997 market estimates. Document 733-R-99-001. Office of Prevention, Pesticides and Toxic Substances, U.S. Environmental Protection Agency, Washington, D.C.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, and D. Moore. 1999. Short protocols in molecular biology: a compendium of methods from Current Protocols in Molecular Biology, 4th ed. John Wiley & Sons, New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

