Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance
- PMID: 12450822
- PMCID: PMC134417
- DOI: 10.1128/AEM.68.12.6005-6012.2002
Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance
Abstract
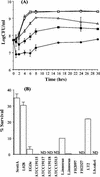
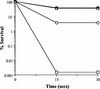
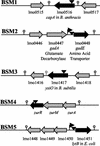
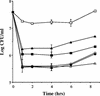
Bile is one of many barriers that Listeria monocytogenes must overcome in the human gastrointestinal tract in order to infect and cause disease. We demonstrated that stationary-phase cultures of L. monocytogenes LO28 were able to tolerate concentrations of bovine, porcine, and human bile and bile acids well in excess of those encountered in vivo. Strain LO28 was relatively bile resistant compared with other clinical isolates of L. monocytogenes, as well as with Listeria innocua, Salmonella enterica serovar Typhimurium LT2, and Lactobacillus sakei. While exponential-phase L. monocytogenes LO28 cells were exquisitely sensitive to unconjugated bile acids, prior adaptation to sublethal levels of bile acids or heterologous stresses, such as acid, heat, salt, or sodium dodecyl sulfate (SDS), significantly enhanced bile resistance. This adaptive response was independent of protein synthesis, and in the cases of bile and SDS adaptation, occurred in seconds. In order to identify genetic loci involved in the bile tolerance phenotype of L. monocytogenes LO28, transposon (Tn917) and plasmid (pORI19) integration banks were screened for bile-sensitive mutants. The disrupted genes included a homologue of the capA locus required for capsule formation in Bacillus anthracis; a gene encoding the transcriptional regulator ZurR; a homologue of an Escherichia coli gene, lytB, involved in isoprenoid biosynthesis; a gene encoding a homologue of the Bacillus subtilis membrane protein YxiO; and a gene encoding an amino acid transporter with a putative role in pH homeostasis, gadE. Interestingly, all of the identified loci play putative roles in maintenance of the cell envelope or in stress responses.
Figures

References
-
- Allerberger, F., B. Langer, O. Hirsch, M. P. Dierich, and H. P. Seeliger. 1989. Listeria monocytogenes cholecystitis. Z. Gastroenterol. 27:145-147. - PubMed
-
- Briones, V., M. M. Blanco, A. Marco, N. Prats, J. F. Fernandez-Garayzabal, G. Suarez, M. Domingo, and L. Dominguez. 1992. Biliary excretion as possible origin of Listeria monocytogenes in fecal carriers. Am. J. Vet. Res. 53:191-193. - PubMed
-
- Carey, M. C., D. M. Small, and C. M. Bliss. 1983. Lipid digestion and absorption. Annu. Rev. Physiol. 45:651-677. - PubMed
-
- Chowdhury, R., G. K. Sahu, and J. Das. 1996. Stress response in pathogenic bacteria. J. Biosci. 21:149-160.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

