The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria)
- PMID: 12450827
- PMCID: PMC134415
- DOI: 10.1128/AEM.68.12.6043-6050.2002
The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria)
Abstract
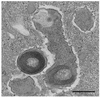
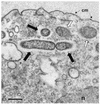
Obligate bacterial endosymbionts of paramecia able to form refractile inclusion bodies (R bodies), thereby conferring a killer trait upon their ciliate hosts, have traditionally been grouped into the genus CAEDIBACTER: Of the six species described to date, only the Paramecium caudatum symbiont Caedibacter caryophilus has been phylogenetically characterized by its 16S rRNA gene sequence, and it was found to be a member of the Alphaproteobacteria related to the RICKETTSIALES: In this study, the Caedibacter taeniospiralis type strain, an R-body-producing cytoplasmatic symbiont of Paramecium tetraurelia strain 51k, was investigated by comparative 16S rRNA sequence analysis and fluorescence in situ hybridization with specific oligonucleotide probes. C. taeniospiralis is not closely related to C. caryophilus (80% 16S rRNA sequence similarity) but forms a novel evolutionary lineage within the Gammaproteobacteria with the family Francisellaceae as a sister group (87% 16S rRNA sequence similarity). These findings demonstrate that the genus Caedibacter is polyphyletic and comprises at least two phylogenetically different bacterial species belonging to two different classes of the PROTEOBACTERIA: Comparative phylogenetic analysis of C. caryophilus, five closely related Acanthamoeba endosymbionts (including one previously uncharacterized amoebal symbiont identified in this study), and their hosts suggests that the progenitor of the alphaproteobacterial C. caryophilus lived within acanthamoebae prior to the infection of paramecia.
Figures





References
-
- Amann, R., N. Springer, W. Ludwig, H. D. Görtz, and K. H. Schleifer. 1991. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351:161-164. - PubMed
-
- Beale, G. H., and A. Jurand. 1966. Three different types of mate-killer (mu) particles in Paramecium aurelia, stock 540. J. Gen. Microbiol. 23:243-252. - PubMed
-
- Beale, G. H., A. Jurand, and J. R. Preer. 1969. The classes of endosymbionts of Paramecium aurelia. J. Cell Sci. 5:65-91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

