Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils
- PMID: 12450836
- PMCID: PMC134418
- DOI: 10.1128/AEM.68.12.6121-6128.2002
Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils
Abstract
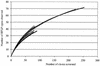
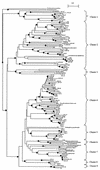
Microorganisms that use nitrate as an alternative terminal electron acceptor play an important role in the global nitrogen cycle. The diversity of the nitrate-reducing community in soil and the influence of the maize roots on the structure of this community were studied. The narG gene encoding the membrane bound nitrate reductase was selected as a functional marker for the nitrate-reducing community. The use of narG is of special interest because the phylogeny of the narG gene closely reflects the 16S ribosomal DNA phylogeny. Therefore, targeting the narG gene provided for the first time a unique insight into the taxonomic composition of the nitrate-reducing community in planted and unplanted soils. The PCR-amplified narG fragments were cloned and analyzed by restriction fragment length polymorphism (RFLP). In all, 60 RFLP types represented by two or more clones were identified in addition to the 58 RFLP types represented by only one clone. At least one clone belonging to each RFLP type was then sequenced. Several of the obtained sequences were not related to the narG genes from cultivated bacteria, suggesting the existence of unidentified nitrate-reducing bacteria in the studied soil. However, environmental sequences were also related to NarG from many bacterial divisions, i.e., Actinobacteria and alpha, beta, and gamma proteobacteria. The presence of the plant roots resulted in a shift in the structure of the nitrate-reducing community between the unplanted and planted soils. Sequencing of RFLP types dominant in the rhizosphere or present only in the rhizosphere revealed that they are related to NarG from the Actinobacteria in an astonishingly high proportion.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. D. Richardson. 1995. Enzymes and associated electron transports systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. - PubMed
-
- Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. - PMC - PubMed
-
- Brunel, B., J. D. Janse, H. J. Laanbroek, and J. D. Woldendorp. 1992. Effect of transient oxic conditions on the composition of the nitrate-reducing community from the rhizosphere of Typha angustipholia. Microbiol. Ecol. 24:51-61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

